 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Eucgr.J00182.1.p | ||||||||
| Common Name | EUGRSUZ_J00182, LOC104421051 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 405aa MW: 45164.5 Da PI: 8.582 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 100.3 | 1.3e-31 | 81 | 134 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
prlrWtp+LH +F++ave+LGG+e+AtPk +l+lm+vkgL+++hvkSHLQ+YR+
Eucgr.J00182.1.p 81 PRLRWTPDLHLCFLHAVERLGGHERATPKLVLQLMNVKGLSIAHVKSHLQMYRS 134
8****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 3.8E-30 | 77 | 135 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 11.404 | 77 | 137 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 7.35E-15 | 79 | 136 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 4.8E-23 | 81 | 136 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 3.6E-9 | 82 | 133 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 405 aa Download sequence Send to blast |
MKKMRNGEST QLTISSPCNK NRKEQDEDSY EEEEEEEEEV GMFKDGRRSS SNSTVDESGK 60 NDGSKVKAGG SVRKYVRSKM PRLRWTPDLH LCFLHAVERL GGHERATPKL VLQLMNVKGL 120 SIAHVKSHLQ MYRSKKTDDP NQAIKDHGFF GDGGDHHVYN LSQLPLLQTF NHRPSSLRFM 180 DASWRGMHNP NQLPGSPVFE RFKIRHIVAS AAGDINCGIN KRFGPPNRDP IHVGHPSFDG 240 EAATTTTAAT CDHYREDHSK SSTLHKSWQK ISQAQVATPS STAKEPDLLR TIEDIRRREQ 300 ATRACLFDDA CGLSSVQEER SRSVLKRKGL DQNAVVDLDL SLRVARPKKE YMENGNNAVD 360 SDLSLSLSPS SRSLFSKLSR LKEDNGDGGG DLKKQDASTL DLTL* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4r_A | 3e-17 | 82 | 136 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 3e-17 | 82 | 136 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 3e-17 | 82 | 136 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 3e-17 | 82 | 136 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00301 | DAP | Transfer from AT2G40260 | Download |
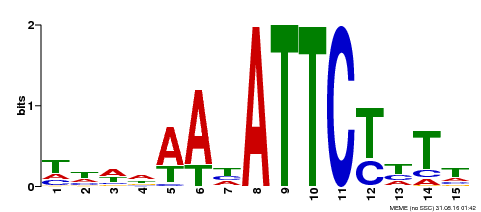
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Eucgr.J00182.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010031180.1 | 0.0 | PREDICTED: myb family transcription factor EFM | ||||
| TrEMBL | A0A059A9S0 | 0.0 | A0A059A9S0_EUCGR; Uncharacterized protein | ||||
| STRING | XP_010031180.1 | 0.0 | (Eucalyptus grandis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM9359 | 21 | 36 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G40260.1 | 2e-47 | G2-like family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Eucgr.J00182.1.p |
| Entrez Gene | 104421051 |



