 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Eucgr.J00491.1.p | ||||||||
| Common Name | EUGRSUZ_J00491, LOC104421301 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 485aa MW: 53622.4 Da PI: 8.3448 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 33.2 | 1.1e-10 | 182 | 223 | 4 | 45 |
XCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 4 lkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaL 45
k rr+++NReAAr+sR+RKka++++Le +L++ ++L
Eucgr.J00491.1.p 182 AKTLRRLAQNREAARKSRLRKKAYVQQLESSRIKLTQLEQDL 223
6889*************************9777777666655 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 4.5E-7 | 179 | 242 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 9.186 | 181 | 225 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 7.4E-8 | 182 | 224 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 5.27E-7 | 183 | 226 | No hit | No description |
| CDD | cd14708 | 2.15E-22 | 183 | 234 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 2.1E-7 | 184 | 227 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 186 | 201 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF14144 | 2.6E-30 | 262 | 337 | IPR025422 | Transcription factor TGA like domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 485 aa Download sequence Send to blast |
MASHRIGEIG LSETGPSTHH VPYSVYHGIN NPTTFINQEG SAFDFGELEE AIVLQGVNIR 60 NDEAKAPLYA GRPAATLDMF PSWPIKFQHT PRGSSKSRGE STDSGSGVNT NTLSSSIAEA 120 RREADSPVSV KGSSSLDHQK LQKPMEMAAS DTSPLPSPSQ SASKAPQDKR KGGSTSEKQL 180 DAKTLRRLAQ NREAARKSRL RKKAYVQQLE SSRIKLTQLE QDLQRARSQG FLLGACGGAN 240 ISSGATIFDM EYARWLEDNH RHMLELRTGL QAHLPDGDLR VIVDGYLSHY DEIFRLKGVA 300 AKSDVFHLIT GVWATPAERC FLWMGGFRPS ELIKILITQL DPLTEQQFMG ICSLQHSSQQ 360 AEEALSQGLE QLQQSLVDTI AGGPSIEGMQ QMAIALGKLT NLEGFVRQAD NLRQHTLHHL 420 RRILTVRQAA RCFLVIGEYY GRLRALSSLW ASRPRESMMN DDNSRQTTTD LQIVQATQNH 480 FSNF* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Together with TGA10, basic leucine-zipper transcription factor required for anther development, probably via the activation of SPL expression in anthers and via the regulation of genes with functions in early and middle tapetal development (PubMed:20805327). Required for signaling responses to pathogen-associated molecular patterns (PAMPs) such as flg22 that involves chloroplastic reactive oxygen species (ROS) production and subsequent expression of H(2)O(2)-responsive genes (PubMed:27717447). {ECO:0000269|PubMed:20805327, ECO:0000269|PubMed:27717447}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00131 | DAP | Transfer from AT1G08320 | Download |
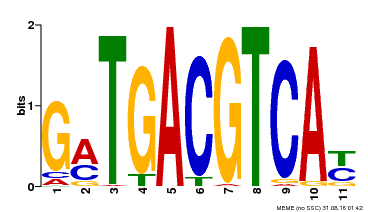
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Eucgr.J00491.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by flg22 in leaves. {ECO:0000269|PubMed:27717447}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010031514.1 | 0.0 | PREDICTED: transcription factor TGA2.2 isoform X1 | ||||
| Swissprot | Q93XM6 | 0.0 | TGA9_ARATH; Transcription factor TGA9 | ||||
| TrEMBL | A0A059A9Q0 | 0.0 | A0A059A9Q0_EUCGR; Uncharacterized protein | ||||
| STRING | XP_010031514.1 | 0.0 | (Eucalyptus grandis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM5524 | 24 | 41 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G08320.3 | 0.0 | bZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Eucgr.J00491.1.p |
| Entrez Gene | 104421301 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



