 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Eucgr.J02113.1.p | ||||||||
| Common Name | EUGRSUZ_J02113, LOC104422542 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 484aa MW: 52813.8 Da PI: 5.7592 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 54.3 | 3.4e-17 | 149 | 198 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV++++++grW+++I+d + k+++lg f+ta Aa+a+++a+ k++g
Eucgr.J02113.1.p 149 SQYRGVTFYRRTGRWESHIWD------CgKQVYLGGFDTAHAAARAYDRAAIKFRG 198
78*******************......55************************998 PP
| |||||||
| 2 | AP2 | 41.2 | 4.2e-13 | 241 | 291 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV+ +k grW+A+ + +k+++lg f+++ eAa+a+++a+ k +g
Eucgr.J02113.1.p 241 SKYRGVTLHK-CGRWEARMGQF--L-GKKYIYLGLFDSEVEAARAYDKAAIKCNG 291
89********.7******5553..2.26**********99**********99776 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF54171 | 4.64E-16 | 149 | 207 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 2.2E-9 | 149 | 198 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 3.9E-17 | 150 | 207 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 17.82 | 150 | 206 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 3.22E-11 | 150 | 208 | No hit | No description |
| SMART | SM00380 | 1.4E-31 | 150 | 212 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 2.30E-25 | 241 | 301 | No hit | No description |
| SuperFamily | SSF54171 | 7.85E-17 | 241 | 300 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 1.3E-8 | 241 | 291 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 1.0E-15 | 242 | 299 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 1.8E-32 | 242 | 305 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 15.527 | 242 | 299 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 5.2E-6 | 243 | 254 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 5.2E-6 | 281 | 301 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 484 aa Download sequence Send to blast |
MLDLNLNEEL SISESAWDES SVAIVKELPE APGAQMDESG TSNSSIVNAD GCDDESCSTR 60 AADARTFSFG ILKAGEGGDG VADDELGVTR QLFPVREVDA DMEWCGESSS LDKRSDVFLV 120 GACKEKEGPR LAMPQQRRKS RRGPRSRSSQ YRGVTFYRRT GRWESHIWDC GKQVYLGGFD 180 TAHAAARAYD RAAIKFRGLD ADINFNLSDY EDDFKQMKNL SKEEFVHILR RQSTGFSRGS 240 SKYRGVTLHK CGRWEARMGQ FLGKKYIYLG LFDSEVEAAR AYDKAAIKCN GREAVTNFEP 300 STYDGEMIAK ASNENSIYGD HGLDLNLGIS ASSRGMVETL EPSDDMRQGS SLRVGNSAAS 360 WGDPSVEGLS MTSGQPPPWT GVYPSVFPCM ERAKEKICGL GLSQGLATWA TQMHGQVNSA 420 PLPLLSAAAS SGFSMSAAHR SPVIPPPILT DSTSQSNRFT SPSSAVNNIS RGYYQLTPQW 480 PPR* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 135 | 141 | QRRKSRR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor (By similarity). Involved in spikelet transition (Probable). Together with IDS1, controls synergistically inflorescence architecture and floral meristem establishment via the regulation of spatio-temporal expression of B- and E-function floral organ identity genes in the lodicules and of spikelet meristem genes (PubMed:22003982). Prevents lemma and palea elongation as well as grain growth (PubMed:28066457). Regulates the transition from spikelet meristem to floral meristem, spikelet meristem determinancy and the floral organ development (PubMed:17144896). {ECO:0000250|UniProtKB:P47927, ECO:0000269|PubMed:17144896, ECO:0000269|PubMed:22003982, ECO:0000269|PubMed:28066457, ECO:0000305|PubMed:26631749}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00615 | PBM | Transfer from AT2G28550 | Download |
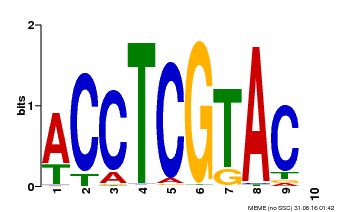
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Eucgr.J02113.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Target of miR172 microRNA mediated cleavage, particularly during floral organ development. {ECO:0000269|PubMed:20017947, ECO:0000269|PubMed:22003982, ECO:0000305|PubMed:26631749, ECO:0000305|PubMed:28066457}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010033195.1 | 0.0 | PREDICTED: floral homeotic protein APETALA 2 isoform X1 | ||||
| Swissprot | Q8H443 | 1e-106 | AP23_ORYSJ; APETALA2-like protein 3 | ||||
| TrEMBL | A0A059AHA6 | 0.0 | A0A059AHA6_EUCGR; Uncharacterized protein | ||||
| STRING | XP_010033195.1 | 0.0 | (Eucalyptus grandis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM5861 | 27 | 47 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G28550.3 | 1e-104 | related to AP2.7 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Eucgr.J02113.1.p |
| Entrez Gene | 104422542 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



