 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Eucgr.J03166.1.p | ||||||||
| Common Name | EUGRSUZ_J03166, LOC104423265 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | GATA | ||||||||
| Protein Properties | Length: 286aa MW: 31048.1 Da PI: 6.5076 | ||||||||
| Description | GATA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GATA | 52.2 | 8.1e-17 | 219 | 255 | 1 | 35 |
GATA 1 CsnCgttk..TplWRrgpdgnktLCnaCGlyyrkkgl 35
C++Cg ++ Tp++Rrgp+g++tLCnaCGl+++ kg+
Eucgr.J03166.1.p 219 CHHCGISEksTPMMRRGPEGPRTLCNACGLMWANKGT 255
*****99999************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00979 | 2.3E-10 | 76 | 111 | IPR010399 | Tify domain |
| PROSITE profile | PS51320 | 14.864 | 76 | 111 | IPR010399 | Tify domain |
| Pfam | PF06200 | 1.0E-12 | 81 | 110 | IPR010399 | Tify domain |
| PROSITE profile | PS51017 | 13.205 | 143 | 185 | IPR010402 | CCT domain |
| Pfam | PF06203 | 1.7E-15 | 143 | 185 | IPR010402 | CCT domain |
| PROSITE profile | PS50114 | 9.011 | 213 | 270 | IPR000679 | Zinc finger, GATA-type |
| SMART | SM00401 | 2.7E-13 | 213 | 266 | IPR000679 | Zinc finger, GATA-type |
| SuperFamily | SSF57716 | 6.65E-13 | 214 | 265 | No hit | No description |
| Gene3D | G3DSA:3.30.50.10 | 5.0E-16 | 218 | 260 | IPR013088 | Zinc finger, NHR/GATA-type |
| PROSITE pattern | PS00344 | 0 | 219 | 246 | IPR000679 | Zinc finger, GATA-type |
| CDD | cd00202 | 3.02E-14 | 219 | 268 | No hit | No description |
| Pfam | PF00320 | 1.1E-14 | 219 | 255 | IPR000679 | Zinc finger, GATA-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 286 aa Download sequence Send to blast |
MDGSHGGDDG MDGGDPTHDR HPMQPHYRHH HGLQAQPHLS NGNDMNDDHD HDGGGGEGVE 60 GDVPSDAGND SDNRGGGDDA DQLTLSFQGQ VYVFDSVPPQ KVQAVLLLLG AREVTPNPPA 120 IPVATNQNQD LMGTPQRSNV PQRLASLIRF REKKKERNFD KKIRYAVRKE VALRMQRNKG 180 QFSSSKPNND ESSSAVTTWE SNQSWGPESN GSPHQEILCH HCGISEKSTP MMRRGPEGPR 240 TLCNACGLMW ANKGTLRDLS KAAHPGVQVS SLNRNEMQSA NLGAD* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that specifically binds 5'-GATA-3' or 5'-GAT-3' motifs within gene promoters. {ECO:0000250, ECO:0000269|PubMed:14966217}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00198 | DAP | Transfer from AT1G51600 | Download |
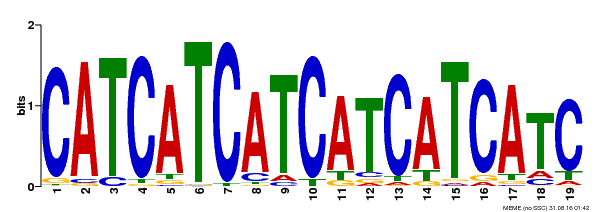
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Eucgr.J03166.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010034076.1 | 0.0 | PREDICTED: GATA transcription factor 28 isoform X1 | ||||
| Swissprot | Q8H1G0 | 1e-110 | GAT28_ARATH; GATA transcription factor 28 | ||||
| TrEMBL | A0A059AJ01 | 0.0 | A0A059AJ01_EUCGR; Uncharacterized protein | ||||
| STRING | XP_010034076.1 | 0.0 | (Eucalyptus grandis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3684 | 26 | 59 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G51600.2 | 7e-90 | ZIM-LIKE 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Eucgr.J03166.1.p |
| Entrez Gene | 104423265 |



