 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | FANhyb_rscf00000124.1.g00015.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Rosoideae; Potentilleae; Fragariinae; Fragaria
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 680aa MW: 75475.1 Da PI: 7.5687 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 52.2 | 1.4e-16 | 85 | 131 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g WT++Ede l +av+++ g++Wk+Ia+ + R++ qc +rwqk+l
FANhyb_rscf00000124.1.g00015.1 85 KGGWTPQEDETLRNAVEKYKGKNWKKIAQNFT-DRSEVQCLHRWQKVL 131
688****************************9.************986 PP
| |||||||
| 2 | Myb_DNA-binding | 61.7 | 1.5e-19 | 137 | 183 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEd+++v++v+++G+ W++I++ ++ gR +kqc++rw+++l
FANhyb_rscf00000124.1.g00015.1 137 KGPWTQEEDDKIVELVAKYGPTKWSLISKSLP-GRIGKQCRERWHNHL 183
79******************************.*************97 PP
| |||||||
| 3 | Myb_DNA-binding | 50.2 | 5.8e-16 | 189 | 232 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
r +WT E++ l +a++++G++ W+ Ia+ ++ gRt++ +k++w++
FANhyb_rscf00000124.1.g00015.1 189 REAWTLVEELALMHAHRMHGNK-WAEIAKALP-GRTDNAIKNHWNS 232
789*******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 15.682 | 80 | 131 | IPR017930 | Myb domain |
| SMART | SM00717 | 9.7E-15 | 84 | 133 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.9E-15 | 85 | 131 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 1.44E-15 | 86 | 141 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 2.6E-18 | 87 | 136 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.87E-13 | 88 | 131 | No hit | No description |
| PROSITE profile | PS51294 | 32.789 | 132 | 187 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 9.36E-31 | 134 | 230 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.7E-18 | 136 | 185 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.1E-18 | 137 | 183 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 8.8E-28 | 137 | 186 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 2.91E-16 | 139 | 183 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 3.1E-22 | 187 | 238 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 6.0E-15 | 188 | 236 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 21.07 | 188 | 238 | IPR017930 | Myb domain |
| Pfam | PF00249 | 1.8E-14 | 189 | 232 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 4.17E-11 | 191 | 234 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 680 aa Download sequence Send to blast |
MSFNLIPSTS SPPEKGRSEV PIEERCLVNK QSTAASSSDS ECSESPILKS PRDWEQPGRG 60 FSIFRTLENP YAGLMRITGP IRRAKGGWTP QEDETLRNAV EKYKGKNWKK IAQNFTDRSE 120 VQCLHRWQKV LNPELVKGPW TQEEDDKIVE LVAKYGPTKW SLISKSLPGR IGKQCRERWH 180 NHLNPDIKRE AWTLVEELAL MHAHRMHGNK WAEIAKALPG RTDNAIKNHW NSSLKKKLDF 240 YLATGQLPPP PKNSYLNVPK ETNRFTATKR LLVGSDKGSD STARTLPGNA DKRRKIDEDG 300 NDHLQSSEPL DMDMCAFYLA NGQLPPVPKN FSPNGAKDTN RTTATTRLLV DSDKGSESSA 360 HTLSGNADTR KVDDNGKDQL QSSGSEDMCG SSSILPDEHA DSEVMERRLR LSLMDPSCSN 420 SESLPTFSNC GVNSKPQIEN SDINSDPKYE NSEFNSQQVE DKVESSTVPV QTPSYGSLYY 480 EPPPVDSSIP FDSGISNLHY LQHEYESSPN LSPISFFTPP TAKSSGLGNR SPESILKIAA 540 KTFPNTPSIL RKRKSAQGHL PPNKLRIVDS ETVRETVTVP EEQERNKNSV ELSGSQDVSL 600 FDSPTSHGIS TTGPNGKAFN ASPPYRLRSR RTAVFKSVEK QLQFTCEEEY AGNSKPVELS 660 ANGSSAVTKD CSHATKMEIT |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h88_C | 8e-65 | 84 | 238 | 5 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 8e-65 | 84 | 238 | 5 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00482 | DAP | Transfer from AT5G02320 | Download |
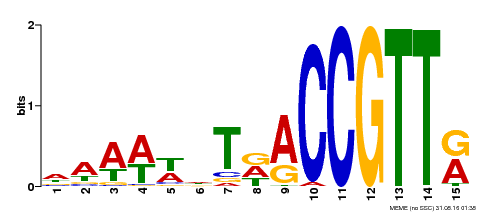
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011466561.1 | 0.0 | PREDICTED: transcriptional activator Myb | ||||
| TrEMBL | A0A2P6RA22 | 0.0 | A0A2P6RA22_ROSCH; Putative transcription factor MYB-HB-like family | ||||
| STRING | XP_004305203.1 | 0.0 | (Fragaria vesca) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF6616 | 34 | 49 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G02320.2 | 1e-114 | myb domain protein 3r-5 | ||||



