 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | FANhyb_rscf00000149.1.g00005.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Rosoideae; Potentilleae; Fragariinae; Fragaria
|
||||||||
| Family | GRAS | ||||||||
| Protein Properties | Length: 606aa MW: 66841.8 Da PI: 5.8478 | ||||||||
| Description | GRAS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GRAS | 453.1 | 1.7e-138 | 238 | 597 | 1 | 374 |
GRAS 1 lvelLlecAeavssgdlelaqalLarlselaspdgdpmqRlaayfteALaarlarsvselykalppsetseknsseela 79
lv+ L++cAeav++++l+la+al++++s la +++ +m+++a++f+eALa+r++r +y a p ++ se l
FANhyb_rscf00000149.1.g00005.1 238 LVHGLMACAEAVQQNNLNLAKALVTQISYLAISQAGAMRKVATFFAEALAQRIFR----VYPAAPIDHS----YSEML- 307
6899***************************************************....6655544444....34444. PP
GRAS 80 alklfsevsPilkfshltaNqaIleavegeervHiiDfdisqGlQWpaLlqaLasRpegppslRiTgvgspesgskeel 158
+ f+e++P+lkf+h+taNqaIle ++g++rvH+iDf+++qG+QWpaL+qaLa Rp+gpp +R+Tg+g+p++++++ l
FANhyb_rscf00000149.1.g00005.1 308 -QMHFYETCPYLKFAHFTANQAILEGFQGKKRVHVIDFSMNQGMQWPALMQALALRPGGPPVFRLTGIGPPAADNSDHL 385
.446*************************************************************************** PP
GRAS 159 eetgerLakfAeelgvpfefnvlvakrledleleeLrvkp..gEalaVnlvlqlhrlldesvsleserdevLklvksls 235
+e+g++La++Ae+++v+fe++ +va++l+dl+ ++L+++p E++aVn+v++lh+ll++++++e+ vL++vk+++
FANhyb_rscf00000149.1.g00005.1 386 QEVGWKLAQLAETIHVEFEYRGFVANSLADLDASMLELRPseVESVAVNSVFELHKLLARPGAIEK----VLSVVKQMK 460
****************************************88899*********************....********* PP
GRAS 236 PkvvvvveqeadhnsesFlerflealeyysalfdsleaklpreseerikvErellgreivnvvacegaerrerhetlek 314
P++v+vveqea+hn++ Fl+rf e+l+yys+lfdsle ++ +++++++++++lg++i nvvaceg er+erhetl++
FANhyb_rscf00000149.1.g00005.1 461 PEIVTVVEQEANHNGPVFLNRFNESLHYYSTLFDSLEGSV---NSQDKMMSEVYLGKQIFNVVACEGVERVERHETLAQ 536
***************************************7...899999999*************************** PP
GRAS 315 WrerleeaGFkpvplsekaakqaklllrkvk.sdgyrveeesgslvlgWkdrpLvsvSaWr 374
Wr r++++GF+pv+l+++a kqa++ll+ ++ +dgyrvee++g+l+lgW++rpL+++SaW+
FANhyb_rscf00000149.1.g00005.1 537 WRTRFDASGFTPVHLGSNAFKQASMLLALFAgGDGYRVEENDGCLMLGWHTRPLIATSAWK 597
************************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF12041 | 1.8E-33 | 43 | 110 | IPR021914 | Transcriptional factor DELLA, N-terminal |
| SMART | SM01129 | 1.1E-32 | 43 | 116 | No hit | No description |
| PROSITE profile | PS50985 | 66.38 | 212 | 576 | IPR005202 | Transcription factor GRAS |
| Pfam | PF03514 | 6.0E-136 | 238 | 597 | IPR005202 | Transcription factor GRAS |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009867 | Biological Process | jasmonic acid mediated signaling pathway | ||||
| GO:0009938 | Biological Process | negative regulation of gibberellic acid mediated signaling pathway | ||||
| GO:0010187 | Biological Process | negative regulation of seed germination | ||||
| GO:0010218 | Biological Process | response to far red light | ||||
| GO:0042176 | Biological Process | regulation of protein catabolic process | ||||
| GO:0042538 | Biological Process | hyperosmotic salinity response | ||||
| GO:2000033 | Biological Process | regulation of seed dormancy process | ||||
| GO:2000377 | Biological Process | regulation of reactive oxygen species metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 606 aa Download sequence Send to blast |
MKREHHSHPH HHPIPNPSIA STSNAKAGKA AMWEESQHDD GMDELLAVLG YKVKSANMAD 60 VAQKIEQLEE FMGTVQQDGL SQLASDTVHY NPSDLSTWLE SMISEITPPP NFEPVMAPPP 120 PLGSSVMMND SFLAPAESST ITSLDFADQS MNTKSKSVSP RTQFEDCSSS NYELKAIPGK 180 AIFSQQTQFD SSPREPKRLK PSPDFYSTTS SSSLPLPSAV ESTRPVVVVD SQENGVRLVH 240 GLMACAEAVQ QNNLNLAKAL VTQISYLAIS QAGAMRKVAT FFAEALAQRI FRVYPAAPID 300 HSYSEMLQMH FYETCPYLKF AHFTANQAIL EGFQGKKRVH VIDFSMNQGM QWPALMQALA 360 LRPGGPPVFR LTGIGPPAAD NSDHLQEVGW KLAQLAETIH VEFEYRGFVA NSLADLDASM 420 LELRPSEVES VAVNSVFELH KLLARPGAIE KVLSVVKQMK PEIVTVVEQE ANHNGPVFLN 480 RFNESLHYYS TLFDSLEGSV NSQDKMMSEV YLGKQIFNVV ACEGVERVER HETLAQWRTR 540 FDASGFTPVH LGSNAFKQAS MLLALFAGGD GYRVEENDGC LMLGWHTRPL IATSAWKLGA 600 QAVMTH |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5b3g_A | 1e-64 | 233 | 596 | 14 | 378 | Protein SCARECROW |
| 5b3h_A | 1e-64 | 233 | 596 | 13 | 377 | Protein SCARECROW |
| 5b3h_D | 1e-64 | 233 | 596 | 13 | 377 | Protein SCARECROW |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcriptional regulator that acts as a repressor of the gibberellin (GA) signaling pathway. Probably acts by participating in large multiprotein complexes that represses transcription of GA-inducible genes. Upon GA application, it is degraded by the proteasome, allowing the GA signaling pathway (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00611 | ChIP-seq | Transfer from AT2G01570 | Download |
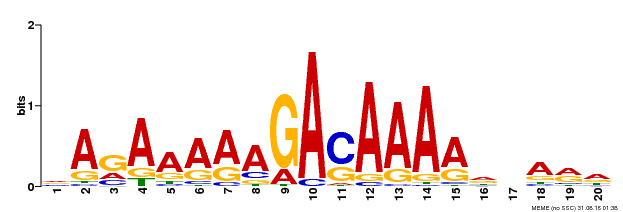
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004298516.1 | 0.0 | PREDICTED: DELLA protein GAIP | ||||
| Swissprot | Q6EI06 | 0.0 | GAIP_CUCMA; DELLA protein GAIP | ||||
| TrEMBL | M9XGL2 | 0.0 | M9XGL2_ROSHC; GAI1 | ||||
| STRING | XP_004298516.1 | 0.0 | (Fragaria vesca) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF803 | 34 | 121 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G01570.1 | 0.0 | GRAS family protein | ||||



