 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | FANhyb_rscf00000584.1.g00004.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Rosoideae; Potentilleae; Fragariinae; Fragaria
|
||||||||
| Family | FAR1 | ||||||||
| Protein Properties | Length: 794aa MW: 90882 Da PI: 6.216 | ||||||||
| Description | FAR1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | FAR1 | 83.7 | 2.9e-26 | 89 | 191 | 1 | 90 |
FAR1 1 kfYneYAkevGFsvrkskskkskrngeitkrtfvCskegkreeekkk.............tekerrtraetrtgCkakl 66
+fY+eYA+++GF++ +++s++sk+++e+++++f Cs++g+++e +k+ e++ +r+ ++t+Cka++
FANhyb_rscf00000584.1.g00004.1 89 SFYQEYARSMGFNTAIQNSRRSKTSREFIDAKFACSRYGTKREYDKTynnkprarqskqdPENATGRRSCSKTDCKASM 167
5******************************************999999999988776555555559999********* PP
FAR1 67 kvkkekdgkwevtkleleHnHela 90
+vk+++dgkw+++++++eHnH l
FANhyb_rscf00000584.1.g00004.1 168 HVKRRSDGKWVIHNFVKEHNHGLL 191
*********************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03101 | 4.2E-24 | 89 | 192 | IPR004330 | FAR1 DNA binding domain |
| Pfam | PF10551 | 8.9E-31 | 290 | 382 | IPR018289 | MULE transposase domain |
| PROSITE profile | PS50966 | 10.151 | 516 | 552 | IPR007527 | Zinc finger, SWIM-type |
| Pfam | PF04434 | 1.1E-6 | 526 | 551 | IPR007527 | Zinc finger, SWIM-type |
| SMART | SM00575 | 1.3E-5 | 527 | 554 | IPR006564 | Zinc finger, PMZ-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009585 | Biological Process | red, far-red light phototransduction | ||||
| GO:0010218 | Biological Process | response to far red light | ||||
| GO:0042753 | Biological Process | positive regulation of circadian rhythm | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 794 aa Download sequence Send to blast |
MDIDLRLPSG DHDKEDEEPN AIDNMLEHEE KMHNGDIESA HIVDDRDEVH AEDGGDLNSP 60 TADMVIFNED TNLEPLFGME FESHGEAYSF YQEYARSMGF NTAIQNSRRS KTSREFIDAK 120 FACSRYGTKR EYDKTYNNKP RARQSKQDPE NATGRRSCSK TDCKASMHVK RRSDGKWVIH 180 NFVKEHNHGL LPAQAVSEQT RKMYAAMARQ FAEYKNVVGL KNDPKNPFDK GRNLSLEAGD 240 LKMLLEFFTH MQSMNSNFFY AIDLGEDQCL KTLLWVDGKS RHDYINFNDV VSLDTTYIRS 300 KYKMPLVLFV GVNQHYQFVL LGCALVSDES ATTFSWLMQT WLKAMGGQAP KVIISDHDQS 360 IKSVVSEVFP NAHHCFCLWN ILGKVSENLG HVIKRHENFL ANFEECIHRP STTEEFEKRW 420 CDIVDEYELK DNDWTQSLYE DRKQWVPTYM TNVCLAGMST VQRSDSVNSF FDKYVHKKTT 480 VQEFLKQYGA ILQDRDRQDE MTTTYRVHDC EKNQDFTVAW NEMKLEVSCS CCLFEYRGYL 540 CRHALIVLQM CQIGTIPDQY ILKRWTKDAK SRHLVVQEPG DVPSRVQKFD DLSQRAMKLI 600 GEGSLSQESY TIVCHALEEA FGNCLAVNNS SKSLVEAGTS GTQGLPCIED DIQNRSIGKG 660 SKKKNPTKKR KVNSEPEVMT VGAQESLQQM DKLNSRAITI DGYYGGQQNV QGMVQLNLMA 720 PTRDNYYGNQ VNQQTIQGLG QLNSIAPSHE SYFSAQQSMH GLGQMEFFRT GFTYIRDDPN 780 VRTAPLHDDA SRHA |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
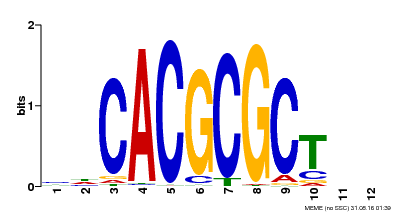
| UniProt | Transcription activator that recognizes and binds to the DNA consensus sequence 5'-CACGCGC-3'. Activates the expression of FHY1 and FHL involved in light responses. When associated with PHYA, protects it from being recognized and degraded by the COP1/SPA complex. Positive regulator of chlorophyll biosynthesis via the activation of HEMB1 gene expression. {ECO:0000269|PubMed:11889039, ECO:0000269|PubMed:12753585, ECO:0000269|PubMed:17012604, ECO:0000269|PubMed:18033885, ECO:0000269|PubMed:18715961, ECO:0000269|PubMed:22634759}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00078 | ChIP-seq | Transfer from AT3G22170 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated after exposure to far-red light. Subject to a negative feedback regulation by PHYA signaling. Up-regulated by white light. {ECO:0000269|PubMed:11889039, ECO:0000269|PubMed:18033885, ECO:0000269|PubMed:22634759}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011467677.1 | 0.0 | PREDICTED: protein FAR-RED ELONGATED HYPOCOTYL 3 | ||||
| Swissprot | Q9LIE5 | 0.0 | FHY3_ARATH; Protein FAR-RED ELONGATED HYPOCOTYL 3 | ||||
| TrEMBL | A0A2P6S0Q3 | 0.0 | A0A2P6S0Q3_ROSCH; Putative transcription factor FAR family | ||||
| STRING | XP_004303947.1 | 0.0 | (Fragaria vesca) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF40 | 30 | 530 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G22170.2 | 0.0 | far-red elongated hypocotyls 3 | ||||



