 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | FANhyb_rscf00000701.1.g00002.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Rosoideae; Potentilleae; Fragariinae; Fragaria
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 731aa MW: 78925.5 Da PI: 6.0732 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 101.7 | 4.1e-32 | 304 | 361 | 2 | 60 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS-- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhek 60
+DgynWrKYGqK+vkgse+prsYY+Ct+++C+vkkkvers+ +++++ei+Y+g Hnh+k
FANhyb_rscf00000701.1.g00002.1 304 EDGYNWRKYGQKQVKGSEYPRSYYKCTHPNCQVKKKVERSH-EGHITEIIYKGAHNHPK 361
7****************************************.***************85 PP
| |||||||
| 2 | WRKY | 102.8 | 1.9e-32 | 520 | 578 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+vkg+++prsYY+Ct agC+v+k+ver+++d k v++tYeg+Hnh+
FANhyb_rscf00000701.1.g00002.1 520 LDDGYRWRKYGQKVVKGNPNPRSYYKCTNAGCTVRKHVERASHDLKSVITTYEGKHNHD 578
59********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF118290 | 1.24E-25 | 301 | 362 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 1.5E-28 | 301 | 362 | IPR003657 | WRKY domain |
| SMART | SM00774 | 6.4E-36 | 303 | 361 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.1E-24 | 304 | 360 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 23.158 | 304 | 362 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 9.3E-37 | 505 | 580 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.96E-28 | 512 | 580 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 38.32 | 515 | 580 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.9E-37 | 520 | 579 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 7.5E-25 | 521 | 578 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009555 | Biological Process | pollen development | ||||
| GO:0009942 | Biological Process | longitudinal axis specification | ||||
| GO:0030010 | Biological Process | establishment of cell polarity | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 731 aa Download sequence Send to blast |
MDDNVAIIGD WLPPSPSPRD FFSSMLVDGI GSRSFLDSPN SNKAEDFFLG SREGSTSGKN 60 LSQGNGTGDE ITEMGSFSEY KSNTRGGLVE RIAARAGFNA PRLNTESIRS SDLSLSSDVR 120 SPYLTIPPGL SPTTLLDSPV FLSNSLAQPS PTTGKYPFVS NGHSRNSTLM TEAADKANFF 180 EDLNSSFAFK PLGESGSFFL GPTSKQTFPS IEVSVQSENS SQNMEPTKVQ NQNANNFQLQ 240 PDFSRTSTEK DNGANTVSAD PRGFDTVGGS TEHSPPPDEQ PEEEGDQRGG GDSMAATGGG 300 TPSEDGYNWR KYGQKQVKGS EYPRSYYKCT HPNCQVKKKV ERSHEGHITE IIYKGAHNHP 360 KPPPNRRSGA IGSSNPLIDM RSDVLEQGGP QTGADGDLVW ASAQKATGGA PDWKHDNLEV 420 TSSASAGPDY GQQSTSMQAQ NGAHLESGDV VDASSTFSND EDEDDRGTHG SVSLVYDGEG 480 DESESKRRKI EAYATEMSGA TRAIREPRVV VQTTSEVDIL DDGYRWRKYG QKVVKGNPNP 540 RSYYKCTNAG CTVRKHVERA SHDLKSVITT YEGKHNHDVP AARNSSHVNS GPSGNMSGQV 600 SGSATQTHPH RPEPSQVHNS MARFERPMSM GSFSLPGRQQ LGPPQGFSFG MNQPGLANLA 660 MAGFGPGQHK VPVLPVHHPY FAQQRQVSEM GFMLPKGEPK VEPMSEPGLN MSNASSVYQQ 720 LMSRLPLGPQ M |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 2e-36 | 304 | 581 | 8 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 2e-36 | 304 | 581 | 8 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Regulates WOX8 and WOX9 expression and basal cell division patterns during early embryogenesis. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element. Required to repolarize the zygote from a transient symmetric state. {ECO:0000269|PubMed:21316593}. | |||||
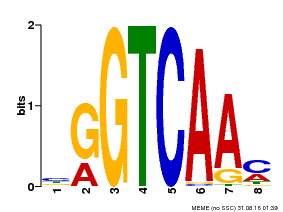
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00071 | PBM | Transfer from AT5G56270 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004294869.1 | 0.0 | PREDICTED: probable WRKY transcription factor 2 | ||||
| Refseq | XP_011461536.1 | 0.0 | PREDICTED: probable WRKY transcription factor 2 | ||||
| Refseq | XP_011461537.1 | 0.0 | PREDICTED: probable WRKY transcription factor 2 | ||||
| Swissprot | Q9FG77 | 0.0 | WRKY2_ARATH; Probable WRKY transcription factor 2 | ||||
| TrEMBL | A0A2P6QL73 | 0.0 | A0A2P6QL73_ROSCH; Putative transcription factor WRKY family | ||||
| STRING | XP_004294869.1 | 0.0 | (Fragaria vesca) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF9524 | 34 | 43 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G56270.1 | 0.0 | WRKY DNA-binding protein 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



