 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GRMZM2G003514_P01 | ||||||||
| Common Name | MADS5, ZEAMMB73_316255 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Andropogonodae; Andropogoneae; Tripsacinae; Zea
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 255aa MW: 28913.8 Da PI: 8.2893 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 97.8 | 4.4e-31 | 9 | 58 | 1 | 50 |
S---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE CS
SRF-TF 1 krienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklyeys 50
krienk+nrqvtfskRrng+lKKA+ELSvLCdaeva+iifs +gklye+
GRMZM2G003514_P01 9 KRIENKINRQVTFSKRRNGLLKKAYELSVLCDAEVALIIFSGRGKLYEFG 58
79**********************************************95 PP
| |||||||
| 2 | K-box | 102.5 | 5.5e-34 | 76 | 172 | 3 | 99 |
K-box 3 kssgksleeakaeslqqelakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkekelqeenkaL 94
++++++ + ++++s++qe++kL+++ e Lqr+qRhllGedL++Ls+keLqqLe+qLe +l++ R++K++l++eq+eel++ e+ l e n++L
GRMZM2G003514_P01 76 AQDSNNGALSESQSWYQEISKLRAKFEALQRTQRHLLGEDLGPLSVKELQQLEKQLECALSQARQRKTQLMMEQVEELRRTERHLGEMNRQL 167
455555677788******************************************************************************** PP
K-box 95 rkkle 99
++kle
GRMZM2G003514_P01 168 KHKLE 172
***97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50066 | 33.235 | 1 | 61 | IPR002100 | Transcription factor, MADS-box |
| SMART | SM00432 | 1.4E-40 | 1 | 60 | IPR002100 | Transcription factor, MADS-box |
| SuperFamily | SSF55455 | 5.23E-33 | 2 | 77 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 5.88E-46 | 2 | 78 | No hit | No description |
| PRINTS | PR00404 | 2.3E-32 | 3 | 23 | IPR002100 | Transcription factor, MADS-box |
| PROSITE pattern | PS00350 | 0 | 3 | 57 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 6.4E-27 | 10 | 57 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 2.3E-32 | 23 | 38 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 2.3E-32 | 38 | 59 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF01486 | 3.1E-30 | 86 | 171 | IPR002487 | Transcription factor, K-box |
| PROSITE profile | PS51297 | 17.377 | 87 | 177 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009911 | Biological Process | positive regulation of flower development | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0048437 | Biological Process | floral organ development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 255 aa Download sequence Send to blast |
MGRGRVELKR IENKINRQVT FSKRRNGLLK KAYELSVLCD AEVALIIFSG RGKLYEFGSA 60 GVTKTLERYQ HCCYNAQDSN NGALSESQSW YQEISKLRAK FEALQRTQRH LLGEDLGPLS 120 VKELQQLEKQ LECALSQARQ RKTQLMMEQV EELRRTERHL GEMNRQLKHK LEAEGCSNYT 180 TLQHAACWPA PGGTIVEHDG ATYQVHPPAH SVAMDCEPTL QIGYPHHQFP PPEAANNIPR 240 SAATGENNFM LGWVL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ox0_A | 1e-21 | 83 | 175 | 13 | 104 | Developmental protein SEPALLATA 3 |
| 4ox0_B | 1e-21 | 83 | 175 | 13 | 104 | Developmental protein SEPALLATA 3 |
| 4ox0_C | 1e-21 | 83 | 175 | 13 | 104 | Developmental protein SEPALLATA 3 |
| 4ox0_D | 1e-21 | 83 | 175 | 13 | 104 | Developmental protein SEPALLATA 3 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Zm.293 | 0.0 | ear | ||||
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | |||||
| Expression Atlas | GRMZM2G003514 | |||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Expressed during ovule development in the inner and outer integuments. Not detected in young panicles. {ECO:0000269|PubMed:12395189, ECO:0000269|PubMed:19820190}. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in the floral meristem. Highly expressed in lodicules. Expressed in palea and pistil. Weakly expressed in carpels, empty glumes and stamens. Not detected in lemmas. {ECO:0000269|PubMed:10444103, ECO:0000269|PubMed:19820190}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. Regulates floral organ identity and floral meristem determinacy. May be involved in the control of flowering time. {ECO:0000269|PubMed:19820190, ECO:0000269|Ref.8}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00315 | DAP | Transfer from AT2G45650 | Download |
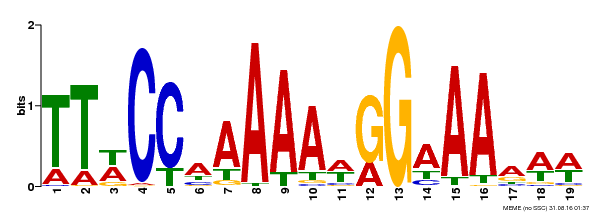
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | GRMZM2G003514_P01 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT087751 | 0.0 | BT087751.1 Zea mays full-length cDNA clone ZM_BFb0184N10 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001105692.2 | 0.0 | MADS box protein | ||||
| Swissprot | Q6EU39 | 1e-144 | MADS6_ORYSJ; MADS-box transcription factor 6 | ||||
| TrEMBL | C4JAA4 | 0.0 | C4JAA4_MAIZE; Agamous-like MADS-box protein AGL6 | ||||
| STRING | GRMZM2G003514_P01 | 0.0 | (Zea mays) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP4187 | 35 | 66 | Representative plant | OGRP16 | 17 | 761 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G45650.1 | 1e-100 | AGAMOUS-like 6 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | GRMZM2G003514_P01 |



