 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GRMZM2G034840_P01 | ||||||||
| Common Name | ARF4 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Andropogonodae; Andropogoneae; Tripsacinae; Zea
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 935aa MW: 102873 Da PI: 6.1207 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 69.4 | 4.9e-22 | 144 | 244 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-S CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldg 86
f+k lt sd++++g +++p++ ae+ ++++++ ++l+++d++ + W++++i ++++r++lt+GW+ Fv a++Lk+gD+v+F +
GRMZM2G034840_P01 144 FCKNLTASDTSTHGGFSVPRRAAEKLfpqldySMQPPN-QELIVRDLHDNMWTFRHI-YRQPKRHLLTTGWSLFVGAKRLKAGDSVLFI--R 231
89*********************999*****9555555.49***************7.5667799************************..4 PP
SSEE..EEEEE-S CS
B3 87 rsefelvvkvfrk 99
+++ +l+v+v+r+
GRMZM2G034840_P01 232 DEKSQLLVGVRRA 244
577778*****97 PP
| |||||||
| 2 | Auxin_resp | 109.5 | 3.5e-36 | 269 | 351 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalkvkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsLk 83
aahaas++ F+v+YnPr+s+s Fv+++++++ a++ + svGmRf m+fete+ss+rr++Gt+vg+sd++p+rWpnSkWr+L+
GRMZM2G034840_P01 269 AAHAASSGGSFTVYYNPRTSPSPFVIPLARYNMATYLQPSVGMRFAMMFETEESSKRRCTGTIVGISDYEPMRWPNSKWRNLQ 351
79*******************************************************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101936 | 3.79E-42 | 134 | 272 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 6.8E-39 | 137 | 257 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 1.81E-19 | 143 | 243 | No hit | No description |
| PROSITE profile | PS50863 | 12.901 | 144 | 245 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 1.5E-19 | 144 | 244 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 1.0E-20 | 144 | 245 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 3.3E-32 | 269 | 351 | IPR010525 | Auxin response factor |
| PROSITE profile | PS51745 | 23.2 | 832 | 916 | IPR000270 | PB1 domain |
| Pfam | PF02309 | 1.5E-8 | 843 | 919 | IPR033389 | AUX/IAA domain |
| SuperFamily | SSF54277 | 4.84E-8 | 851 | 911 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0009908 | Biological Process | flower development | ||||
| GO:0009942 | Biological Process | longitudinal axis specification | ||||
| GO:0010305 | Biological Process | leaf vascular tissue pattern formation | ||||
| GO:0048364 | Biological Process | root development | ||||
| GO:0048507 | Biological Process | meristem development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0016020 | Cellular Component | membrane | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 935 aa Download sequence Send to blast |
MMTSSYEKAT SGVLRNAAAL LDEMQLMGET QGAKKVINSE LWHACAGPLV CLPQRGSLVY 60 YFPQGHSEQV AATTKKIPNS RIPNYPSLPS QLLCQVHNIT LHADKETDEI YAQMTLQPVH 120 SETDVFPIPT LGAYTKSKHP SEYFCKNLTA SDTSTHGGFS VPRRAAEKLF PQLDYSMQPP 180 NQELIVRDLH DNMWTFRHIY RQPKRHLLTT GWSLFVGAKR LKAGDSVLFI RDEKSQLLVG 240 VRRATRQQPA LSSSVLSTDS MHIGVLAAAA HAASSGGSFT VYYNPRTSPS PFVIPLARYN 300 MATYLQPSVG MRFAMMFETE ESSKRRCTGT IVGISDYEPM RWPNSKWRNL QVEWDEHGYG 360 ERPERVSLWD IETPENMVFS SPLNSKRQCL PSYGVSGLHV SSISKPQGSP FGNLQHMPGI 420 SSDIALLLLN QSAQNLGSSI ACQQSSFSSI IQNAKQSYFP PTTLGASTGW NESQQQLNAL 480 GIQKGDQVSC DVQPGIDSIT APEMNVKPRV PRSTDSYSSQ SISDPNSKSD PKTKTRRSKK 540 SSSHKTISDK SEISSVPSQI CDKQRHGSEP TSADFEAEQA TCGNNEDSSG ALTRGDFAGE 600 LQVQQVEQDG LLPPPKLESS KSPDGGKSVS SFPNQGCFSQ FFEGLDWMIP PSCYQDSNGI 660 HSVTTSDSIF NPSEGIPPST MNADGMDAFQ TSCLSECFPN SIQEFISSPD INTLTFMSPE 720 MQHLDAQHDG SNLQSTSNSY VQMSFSEESQ SASLSGLHME AVHINSSCLQ PLATGSFDAG 780 TFSKLSNIKE CQALPLQEIH NSSMGTPSCS MDAAAVEYCM DRSVKPLKPP VRTYTKVQKL 840 GSVGRSIDVT RFRDYHELRS AIACMFGLQG KLEHPGSSDW KLVYVDYEND VLLVGDDPWE 900 EFINCVRCIR ILAPSEVQQM SENGVHVLND CMQMA |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldu_A | 0.0 | 18 | 374 | 31 | 390 | Auxin response factor 5 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Zm.24371 | 0.0 | ear| meristem| shoot | ||||
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | |||||
| Expression Atlas | GRMZM2G034840 | |||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in roots, culms, leaves and young panicles. {ECO:0000269|PubMed:17408882}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00153 | DAP | Transfer from AT1G19850 | Download |
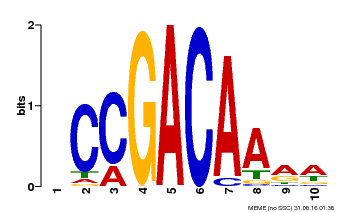
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | GRMZM2G034840_P01 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT066632 | 0.0 | BT066632.1 Zea mays full-length cDNA clone ZM_BFb0041D15 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008668391.1 | 0.0 | auxin response factor 4 isoform X2 | ||||
| Swissprot | Q01K26 | 0.0 | ARFK_ORYSI; Auxin response factor 11 | ||||
| Swissprot | Q8S983 | 0.0 | ARFK_ORYSJ; Auxin response factor 11 | ||||
| TrEMBL | A0A1D6DUQ8 | 0.0 | A0A1D6DUQ8_MAIZE; Auxin response factor | ||||
| STRING | GRMZM2G034840_P02 | 0.0 | (Zea mays) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G19850.1 | 0.0 | ARF family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | GRMZM2G034840_P01 |
| Entrez Gene | 100383226 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



