 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GRMZM2G056600_P01 | ||||||||
| Common Name | cl52289_1b | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Andropogonodae; Andropogoneae; Tripsacinae; Zea
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 261aa MW: 28461.5 Da PI: 4.5039 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 60.5 | 2.7e-19 | 48 | 101 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k++++++eq+++Le+ Fe ++++ e++++LA+ lgL+ rqV vWFqNrRa++k
GRMZM2G056600_P01 48 KKRRLSSEQVRALERSFEVENKLEPERKARLARDLGLQPRQVAVWFQNRRARWK 101
45689************************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 134.4 | 3.9e-43 | 47 | 139 | 1 | 93 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreelk 92
ekkrrls+eqv++LE+sFe e+kLeperK++lar+LglqprqvavWFqnrRAR+ktkqlE+dy+aL+++ydal++++++L++++++L +e+k
GRMZM2G056600_P01 47 EKKRRLSSEQVRALERSFEVENKLEPERKARLARDLGLQPRQVAVWFQNRRARWKTKQLERDYSALRQSYDALRHDHDALRRDKDALLAEIK 138
69**************************************************************************************9998 PP
HD-ZIP_I/II 93 e 93
e
GRMZM2G056600_P01 139 E 139
7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 4.6E-20 | 36 | 105 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.572 | 43 | 103 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.0E-18 | 46 | 107 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 7.48E-19 | 48 | 104 | No hit | No description |
| Pfam | PF00046 | 1.1E-16 | 48 | 101 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 3.4E-20 | 49 | 110 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 8.6E-6 | 74 | 83 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 78 | 101 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 8.6E-6 | 83 | 99 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 2.2E-17 | 103 | 144 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009637 | Biological Process | response to blue light | ||||
| GO:0030308 | Biological Process | negative regulation of cell growth | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0048510 | Biological Process | regulation of timing of transition from vegetative to reproductive phase | ||||
| GO:0048573 | Biological Process | photoperiodism, flowering | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Plant Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PO Term | PO Category | PO Description | ||||
| PO:0000037 | anatomy | shoot apex | ||||
| PO:0006310 | anatomy | tassel floret | ||||
| PO:0006339 | anatomy | juvenile vascular leaf | ||||
| PO:0006340 | anatomy | adult vascular leaf | ||||
| PO:0006341 | anatomy | primary shoot system | ||||
| PO:0006354 | anatomy | ear floret | ||||
| PO:0006505 | anatomy | central spike of ear inflorescence | ||||
| PO:0008018 | anatomy | transition vascular leaf | ||||
| PO:0009001 | anatomy | fruit | ||||
| PO:0009009 | anatomy | plant embryo | ||||
| PO:0009025 | anatomy | vascular leaf | ||||
| PO:0009054 | anatomy | inflorescence bract | ||||
| PO:0009066 | anatomy | anther | ||||
| PO:0009074 | anatomy | style | ||||
| PO:0009084 | anatomy | pericarp | ||||
| PO:0009089 | anatomy | endosperm | ||||
| PO:0020040 | anatomy | leaf base | ||||
| PO:0020104 | anatomy | leaf sheath | ||||
| PO:0020126 | anatomy | tassel inflorescence | ||||
| PO:0020127 | anatomy | primary root | ||||
| PO:0020136 | anatomy | ear inflorescence | ||||
| PO:0020142 | anatomy | stem internode | ||||
| PO:0020148 | anatomy | shoot apical meristem | ||||
| PO:0025142 | anatomy | leaf tip | ||||
| PO:0025287 | anatomy | seedling coleoptile | ||||
| PO:0001007 | developmental stage | pollen development stage | ||||
| PO:0001009 | developmental stage | D pollen mother cell meiosis stage | ||||
| PO:0001052 | developmental stage | vascular leaf expansion stage | ||||
| PO:0001053 | developmental stage | vascular leaf post-expansion stage | ||||
| PO:0001083 | developmental stage | inflorescence development stage | ||||
| PO:0001094 | developmental stage | plant embryo coleoptilar stage | ||||
| PO:0001095 | developmental stage | plant embryo true leaf formation stage | ||||
| PO:0001180 | developmental stage | plant proembryo stage | ||||
| PO:0007001 | developmental stage | early whole plant fruit ripening stage | ||||
| PO:0007003 | developmental stage | IL.03 full inflorescence length reached stage | ||||
| PO:0007006 | developmental stage | IL.00 inflorescence just visible stage | ||||
| PO:0007015 | developmental stage | radicle emergence stage | ||||
| PO:0007016 | developmental stage | whole plant flowering stage | ||||
| PO:0007022 | developmental stage | seed imbibition stage | ||||
| PO:0007026 | developmental stage | FL.00 first flower(s) open stage | ||||
| PO:0007031 | developmental stage | mid whole plant fruit ripening stage | ||||
| PO:0007032 | developmental stage | whole plant fruit formation stage up to 10% | ||||
| PO:0007045 | developmental stage | coleoptile emergence stage | ||||
| PO:0007063 | developmental stage | LP.07 seven leaves visible stage | ||||
| PO:0007065 | developmental stage | LP.05 five leaves visible stage | ||||
| PO:0007072 | developmental stage | LP.18 eighteen leaves visible stage | ||||
| PO:0007094 | developmental stage | LP.01 one leaf visible stage | ||||
| PO:0007101 | developmental stage | LP.09 nine leaves visible stage | ||||
| PO:0007104 | developmental stage | LP.15 fifteen leaves visible stage | ||||
| PO:0007106 | developmental stage | LP.03 three leaves visible stage | ||||
| PO:0007112 | developmental stage | 1 main shoot growth stage | ||||
| PO:0007116 | developmental stage | LP.11 eleven leaves visible stage | ||||
| PO:0007123 | developmental stage | LP.06 six leaves visible stage | ||||
| PO:0007633 | developmental stage | endosperm development stage | ||||
| PO:0021004 | developmental stage | inflorescence initiation stage | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 261 aa Download sequence Send to blast |
MKRPRGASPP LAAMTNPNDD GYGVVGMEAD ADADADEEMM ACGGGGEKKR RLSSEQVRAL 60 ERSFEVENKL EPERKARLAR DLGLQPRQVA VWFQNRRARW KTKQLERDYS ALRQSYDALR 120 HDHDALRRDK DALLAEIKEL KAKLGDEEAA ASFTSVKAEP AASDGPPPVG VGSSESDSSA 180 VLNDADPPVA EAPVPEVRGT LLDAPGAVAA NHGGVFFHGS FLKVEEDETG LLDDDEPCGG 240 FFSVEQPPPM AWWTEPTEHW N |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 72 | 78 | ERKARLA |
| 2 | 95 | 103 | RRARWKTKQ |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Zm.4511 | 0.0 | ear| endosperm| meristem| ovary| shoot| tassel | ||||
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | |||||
| Expression Atlas | GRMZM2G056600 | |||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in leaf and floral organ primordia, floral meristems, embryonic axis and cells surrounding the vascular bundles. Expressed in the vasculature of roots, stem, leaves and spikelets, and in the vascular bundle of the scutellum in embryos. {ECO:0000269|PubMed:10732669, ECO:0000269|PubMed:17999151, ECO:0000269|PubMed:18049796}. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in leaf and floral organ primordia, floral meristems, embryonic axis and cells surrounding the vascular bundles. Expressed in the vasculature of roots, stem, leaves and spikelets, and in the vascular bundle of the scutellum in embryos. {ECO:0000269|PubMed:10732669, ECO:0000269|PubMed:17999151, ECO:0000269|PubMed:18049796}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription activator that binds to the DNA sequence 5'-CAAT[AT]ATTG-3'. May be involved in the regulation of gibberellin signaling. {ECO:0000269|PubMed:10732669, ECO:0000269|PubMed:18049796}. | |||||
| UniProt | Probable transcription activator that binds to the DNA sequence 5'-CAAT[AT]ATTG-3'. May be involved in the regulation of gibberellin signaling. {ECO:0000269|PubMed:10732669, ECO:0000269|PubMed:18049796}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00051 | PBM | Transfer from AT4G40060 | Download |
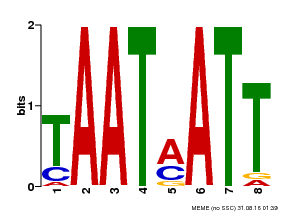
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | GRMZM2G056600_P01 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By gibberellin. Down-regulated in leaves by drought stress. {ECO:0000269|PubMed:17999151, ECO:0000269|PubMed:18049796}. | |||||
| UniProt | INDUCTION: By gibberellin. Down-regulated in leaves by drought stress. {ECO:0000269|PubMed:17999151, ECO:0000269|PubMed:18049796}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT034257 | 0.0 | BT034257.1 Zea mays full-length cDNA clone ZM_BFc0009H14 mRNA, complete cds. | |||
| GenBank | BT039062 | 0.0 | BT039062.1 Zea mays full-length cDNA clone ZM_BFb0367N11 mRNA, complete cds. | |||
| GenBank | BT055374 | 0.0 | BT055374.1 Zea mays full-length cDNA clone ZM_BFb0091J03 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001140555.1 | 0.0 | uncharacterized protein LOC100272620 | ||||
| Swissprot | Q6K498 | 1e-126 | HOX4_ORYSJ; Homeobox-leucine zipper protein HOX4 | ||||
| Swissprot | Q9XH37 | 1e-126 | HOX4_ORYSI; Homeobox-leucine zipper protein HOX4 | ||||
| TrEMBL | B4FAW9 | 0.0 | B4FAW9_MAIZE; Homeobox-leucine zipper protein ATHB-6 | ||||
| TrEMBL | K4JC73 | 0.0 | K4JC73_MAIZE; HB-type transcription factor (Fragment) | ||||
| STRING | GRMZM2G056600_P01 | 0.0 | (Zea mays) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3009 | 34 | 82 | Representative plant | OGRP129 | 16 | 189 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G40060.1 | 2e-44 | homeobox protein 16 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | GRMZM2G056600_P01 |
| Entrez Gene | 100272620 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



