 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GRMZM2G058479_P02 | ||||||||
| Common Name | LOC100277669, ZIM36, Zm.96553 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Andropogonodae; Andropogoneae; Tripsacinae; Zea
|
||||||||
| Family | GATA | ||||||||
| Protein Properties | Length: 253aa MW: 26664.1 Da PI: 9.2473 | ||||||||
| Description | GATA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GATA | 54.5 | 1.6e-17 | 164 | 200 | 1 | 35 |
GATA 1 CsnCgttk..TplWRrgpdgnktLCnaCGlyyrkkgl 35
C++Cgt Tp++Rrgpdg++tLCnaCGl+++ kgl
GRMZM2G058479_P02 164 CHHCGTNAtaTPMMRRGPDGPRTLCNACGLMWANKGL 200
******9989***********************9997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00979 | 9.6E-12 | 29 | 64 | IPR010399 | Tify domain |
| PROSITE profile | PS51320 | 15.373 | 29 | 64 | IPR010399 | Tify domain |
| Pfam | PF06200 | 3.4E-13 | 32 | 63 | IPR010399 | Tify domain |
| Pfam | PF06203 | 5.0E-16 | 88 | 130 | IPR010402 | CCT domain |
| PROSITE profile | PS51017 | 13.404 | 88 | 130 | IPR010402 | CCT domain |
| PROSITE profile | PS50114 | 9.605 | 158 | 215 | IPR000679 | Zinc finger, GATA-type |
| SMART | SM00401 | 1.7E-12 | 158 | 216 | IPR000679 | Zinc finger, GATA-type |
| SuperFamily | SSF57716 | 4.7E-12 | 162 | 211 | No hit | No description |
| CDD | cd00202 | 2.02E-15 | 163 | 206 | No hit | No description |
| Gene3D | G3DSA:3.30.50.10 | 8.0E-16 | 163 | 204 | IPR013088 | Zinc finger, NHR/GATA-type |
| Pfam | PF00320 | 1.8E-15 | 164 | 200 | IPR000679 | Zinc finger, GATA-type |
| PROSITE pattern | PS00344 | 0 | 164 | 191 | IPR000679 | Zinc finger, GATA-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Plant Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PO Term | PO Category | PO Description | ||||
| PO:0000037 | anatomy | shoot apex | ||||
| PO:0006310 | anatomy | tassel floret | ||||
| PO:0006339 | anatomy | juvenile vascular leaf | ||||
| PO:0006340 | anatomy | adult vascular leaf | ||||
| PO:0006341 | anatomy | primary shoot system | ||||
| PO:0006354 | anatomy | ear floret | ||||
| PO:0006505 | anatomy | central spike of ear inflorescence | ||||
| PO:0008018 | anatomy | transition vascular leaf | ||||
| PO:0009001 | anatomy | fruit | ||||
| PO:0009009 | anatomy | plant embryo | ||||
| PO:0009025 | anatomy | vascular leaf | ||||
| PO:0009054 | anatomy | inflorescence bract | ||||
| PO:0009066 | anatomy | anther | ||||
| PO:0009074 | anatomy | style | ||||
| PO:0009084 | anatomy | pericarp | ||||
| PO:0009089 | anatomy | endosperm | ||||
| PO:0020040 | anatomy | leaf base | ||||
| PO:0020104 | anatomy | leaf sheath | ||||
| PO:0020126 | anatomy | tassel inflorescence | ||||
| PO:0020127 | anatomy | primary root | ||||
| PO:0020136 | anatomy | ear inflorescence | ||||
| PO:0020142 | anatomy | stem internode | ||||
| PO:0020148 | anatomy | shoot apical meristem | ||||
| PO:0025142 | anatomy | leaf tip | ||||
| PO:0025287 | anatomy | seedling coleoptile | ||||
| PO:0001007 | developmental stage | pollen development stage | ||||
| PO:0001009 | developmental stage | D pollen mother cell meiosis stage | ||||
| PO:0001052 | developmental stage | vascular leaf expansion stage | ||||
| PO:0001053 | developmental stage | vascular leaf post-expansion stage | ||||
| PO:0001083 | developmental stage | inflorescence development stage | ||||
| PO:0001094 | developmental stage | plant embryo coleoptilar stage | ||||
| PO:0001095 | developmental stage | plant embryo true leaf formation stage | ||||
| PO:0001180 | developmental stage | plant proembryo stage | ||||
| PO:0007001 | developmental stage | early whole plant fruit ripening stage | ||||
| PO:0007003 | developmental stage | IL.03 full inflorescence length reached stage | ||||
| PO:0007006 | developmental stage | IL.00 inflorescence just visible stage | ||||
| PO:0007015 | developmental stage | radicle emergence stage | ||||
| PO:0007016 | developmental stage | whole plant flowering stage | ||||
| PO:0007022 | developmental stage | seed imbibition stage | ||||
| PO:0007026 | developmental stage | FL.00 first flower(s) open stage | ||||
| PO:0007031 | developmental stage | mid whole plant fruit ripening stage | ||||
| PO:0007032 | developmental stage | whole plant fruit formation stage up to 10% | ||||
| PO:0007045 | developmental stage | coleoptile emergence stage | ||||
| PO:0007063 | developmental stage | LP.07 seven leaves visible stage | ||||
| PO:0007065 | developmental stage | LP.05 five leaves visible stage | ||||
| PO:0007072 | developmental stage | LP.18 eighteen leaves visible stage | ||||
| PO:0007094 | developmental stage | LP.01 one leaf visible stage | ||||
| PO:0007101 | developmental stage | LP.09 nine leaves visible stage | ||||
| PO:0007104 | developmental stage | LP.15 fifteen leaves visible stage | ||||
| PO:0007106 | developmental stage | LP.03 three leaves visible stage | ||||
| PO:0007112 | developmental stage | 1 main shoot growth stage | ||||
| PO:0007116 | developmental stage | LP.11 eleven leaves visible stage | ||||
| PO:0007123 | developmental stage | LP.06 six leaves visible stage | ||||
| PO:0007633 | developmental stage | endosperm development stage | ||||
| PO:0021004 | developmental stage | inflorescence initiation stage | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 253 aa Download sequence Send to blast |
MDADAAAAAA AAAVSQMDPH SALVAGTVPP MATNQLTLSF QGEVYVFDSV SPDKVQAVLL 60 LLGGRELSSL GGASSSAPYS KRLNYPHRVA SLMRFREKRK ERNFDKKIRY SVRKEVALRM 120 QRNRGQFTSS KPKPDEIAAS EMASADGSPN WALVEGRPPS AAECHHCGTN ATATPMMRRG 180 PDGPRTLCNA CGLMWANKGL LRDVTKSPVP LQATQSAPHL DGGNGSAMSA PGSELENAAA 240 AMTNGHESSS SGV |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Zm.96553 | 0.0 | cell lignification| ear| endosperm| meristem| ovary| shoot| tassel | ||||
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | |||||
| Expression Atlas | GRMZM2G058479 | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that specifically binds 5'-GATA-3' or 5'-GAT-3' motifs within gene promoters. {ECO:0000250|UniProtKB:Q8LAU9}. | |||||
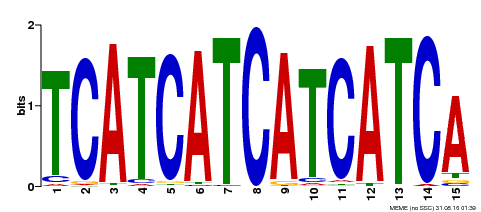
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00369 | DAP | Transfer from AT3G21175 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | GRMZM2G058479_P02 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By abscisic acid (ABA), and drought and salt stresses. Down-regulated by jasmonate and wounding. {ECO:0000269|PubMed:19618278}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EU969898 | 0.0 | EU969898.1 Zea mays clone 337006 hypothetical protein mRNA, complete cds. | |||
| GenBank | KU512872 | 0.0 | KU512872.1 Zea mays TIFY21 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001144648.2 | 0.0 | uncharacterized protein LOC100277669 | ||||
| Swissprot | Q5Z4U5 | 1e-128 | GAT20_ORYSJ; GATA transcription factor 20 | ||||
| TrEMBL | A0A060D777 | 0.0 | A0A060D777_MAIZE; GATA transcription factor 24 (Fragment) | ||||
| TrEMBL | A0A1D6GUV8 | 0.0 | A0A1D6GUV8_MAIZE; GATA transcription factor 24 | ||||
| STRING | GRMZM2G058479_P01 | 0.0 | (Zea mays) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G21175.2 | 1e-65 | ZIM-like 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | GRMZM2G058479_P02 |
| Entrez Gene | 100277669 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



