 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GRMZM2G092517_P01 | ||||||||
| Common Name | LOB22, LOC100285303, Zm.26782 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Andropogonodae; Andropogoneae; Tripsacinae; Zea
|
||||||||
| Family | LBD | ||||||||
| Protein Properties | Length: 237aa MW: 25441.7 Da PI: 6.5074 | ||||||||
| Description | LBD family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF260 | 127.8 | 5.1e-40 | 54 | 153 | 1 | 100 |
DUF260 1 aCaaCkvlrrkCakdCvlapyfpaeqpkkfanvhklFGasnvlkllkalpeeeredamsslvyeAearardPvyGavgvilklqqqleqlka 92
+CaaCk+lrr+Ca++C++apyf ++p+kfa+vhk+FGasnv+k+l ++ e er d++sslvyeA+ r+rdPvyG++g i+ lqqq++ l+a
GRMZM2G092517_P01 54 PCAACKLLRRRCAQECPFAPYFSPHEPHKFAAVHKVFGASNVSKMLLEVVEGERADTASSLVYEANLRLRDPVYGCMGAISILQQQVNALEA 145
7******************************************************************************************* PP
DUF260 93 elallkee 100
el+++++e
GRMZM2G092517_P01 146 ELEAVRAE 153
***99987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50891 | 24.755 | 53 | 154 | IPR004883 | Lateral organ boundaries, LOB |
| Pfam | PF03195 | 9.4E-40 | 54 | 151 | IPR004883 | Lateral organ boundaries, LOB |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0016020 | Cellular Component | membrane | ||||
| Plant Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PO Term | PO Category | PO Description | ||||
| PO:0006340 | anatomy | adult vascular leaf | ||||
| PO:0008018 | anatomy | transition vascular leaf | ||||
| PO:0009009 | anatomy | plant embryo | ||||
| PO:0009025 | anatomy | vascular leaf | ||||
| PO:0020040 | anatomy | leaf base | ||||
| PO:0020127 | anatomy | primary root | ||||
| PO:0001052 | developmental stage | vascular leaf expansion stage | ||||
| PO:0001095 | developmental stage | plant embryo true leaf formation stage | ||||
| PO:0007001 | developmental stage | early whole plant fruit ripening stage | ||||
| PO:0007015 | developmental stage | radicle emergence stage | ||||
| PO:0007031 | developmental stage | mid whole plant fruit ripening stage | ||||
| PO:0007063 | developmental stage | LP.07 seven leaves visible stage | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 237 aa Download sequence Send to blast |
MSTERERLDE IGKIKREPDT AALAVAAASA STVPADNHIP RRLGHGGALN TLTPCAACKL 60 LRRRCAQECP FAPYFSPHEP HKFAAVHKVF GASNVSKMLL EVVEGERADT ASSLVYEANL 120 RLRDPVYGCM GAISILQQQV NALEAELEAV RAEILKHRYR QATGNRLMDD VRATASFVLA 180 PASAPMHVND VVSVVEAGQE VTATGGASGV YIAEAEQPSS TNNYSSLNPS EHPAYFG |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5ly0_A | 3e-41 | 54 | 157 | 11 | 114 | LOB family transfactor Ramosa2.1 |
| 5ly0_B | 3e-41 | 54 | 157 | 11 | 114 | LOB family transfactor Ramosa2.1 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Zm.26782 | 0.0 | meristem | ||||
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | |||||
| Expression Atlas | GRMZM2G092517 | |||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in shoots and roots and at low levels in flowers, but not in leaves or inflorescence stems. {ECO:0000269|PubMed:12068116}. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in young shoots, roots, stems, leaves and flowers. {ECO:0000269|PubMed:12068116}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00283 | DAP | Transfer from AT2G30340 | Download |
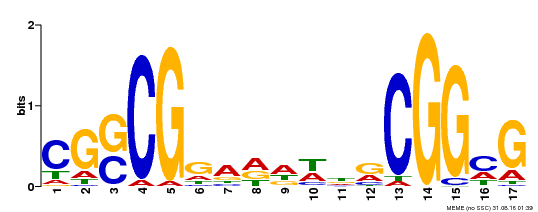
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | GRMZM2G092517_P01 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT068699 | 0.0 | BT068699.1 Zea mays full-length cDNA clone ZM_BFb0375M24 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001151668.2 | 1e-175 | LOB domain protein 25 | ||||
| Swissprot | Q8L5T5 | 8e-67 | LBD15_ARATH; LOB domain-containing protein 15 | ||||
| Swissprot | Q9AT61 | 1e-66 | LBD13_ARATH; LOB domain-containing protein 13 | ||||
| TrEMBL | C0PK80 | 1e-174 | C0PK80_MAIZE; LOB domain-containing protein 13 | ||||
| STRING | GRMZM2G092517_P01 | 1e-175 | (Zea mays) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2630 | 38 | 91 | Representative plant | OGRP60 | 16 | 318 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G40470.1 | 4e-63 | LOB domain-containing protein 15 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | GRMZM2G092517_P01 |
| Entrez Gene | 100285303 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



