 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GRMZM2G139963_P01 | ||||||||
| Common Name | HB102, Zm.83544 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Andropogonodae; Andropogoneae; Tripsacinae; Zea
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 344aa MW: 37604.1 Da PI: 6.7673 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 55.7 | 8.5e-18 | 120 | 173 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k+++++ eq+++Le+ Fe +++ e++ +LA+ lgL+ rqV +WFqNrRa++k
GRMZM2G139963_P01 120 KKRRLNVEQVRTLEKNFELGNKLEPERKLQLARALGLQPRQVAIWFQNRRARWK 173
4568999**********************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 128 | 4e-41 | 119 | 209 | 1 | 91 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreel 91
ekkrrl+ eqv++LE++Fe +kLeperK +lar+Lglqprqva+WFqnrRAR+ktkqlEkdy+aLkr++da+k++n++L +++++L++e+
GRMZM2G139963_P01 119 EKKRRLNVEQVRTLEKNFELGNKLEPERKLQLARALGLQPRQVAIWFQNRRARWKTKQLEKDYDALKRQLDAVKADNDALLSHNKKLQAEI 209
69**************************************************************************************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 2.27E-19 | 111 | 177 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.086 | 115 | 175 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.3E-17 | 118 | 179 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 3.4E-15 | 120 | 173 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 2.49E-17 | 120 | 176 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 2.7E-18 | 122 | 183 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 1.7E-5 | 146 | 155 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 150 | 173 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 1.7E-5 | 155 | 171 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 3.1E-14 | 175 | 215 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009744 | Biological Process | response to sucrose | ||||
| GO:0048826 | Biological Process | cotyledon morphogenesis | ||||
| GO:0080022 | Biological Process | primary root development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 344 aa Download sequence Send to blast |
MRPMASNGMA SAPSPFFPPN FLLQMQQTPS DHEHQEQHHH HHHHKHHLAA PPLHPHHNPF 60 LPSSQCPSLQ DFRGMAPIML GKRPMYGADV QVVGGEEVNG CGGVNEDELS DDGSQAGGEK 120 KRRLNVEQVR TLEKNFELGN KLEPERKLQL ARALGLQPRQ VAIWFQNRRA RWKTKQLEKD 180 YDALKRQLDA VKADNDALLS HNKKLQAEIL SLKGREAGGS SELINLNKET EASCSNRSEN 240 SSEINLDISR APASEAPLDP TPPPGAGGGG MIPFYPPSVG GRPASAAGVD IDQLLHTSAP 300 KLEQHGSGGA VVVQAAETAS FGNLLCGVDE PPPFWPWADH QHFH |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 167 | 175 | RRARWKTKQ |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Zm.83544 | 0.0 | endosperm| meristem | ||||
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | |||||
| Expression Atlas | GRMZM2G139963 | |||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in seedlings, roots, stems, leaf blades and panicles. {ECO:0000269|PubMed:17999151}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
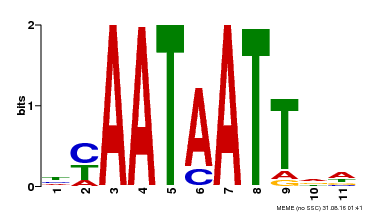
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00225 | DAP | Transfer from AT1G69780 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | GRMZM2G139963_P01 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EU966490 | 0.0 | EU966490.1 Zea mays clone 294849 homeobox-leucine zipper protein HAT7 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001353536.1 | 0.0 | homeobox-leucine zipper protein HAT7 | ||||
| Swissprot | Q8S7W9 | 1e-141 | HOX21_ORYSJ; Homeobox-leucine zipper protein HOX21 | ||||
| TrEMBL | A0A060D3B5 | 0.0 | A0A060D3B5_MAIZE; HB transcription factor (Fragment) | ||||
| STRING | GRMZM2G139963_P01 | 0.0 | (Zea mays) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2289 | 38 | 94 | Representative plant | OGRP129 | 16 | 189 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G69780.1 | 2e-80 | HD-ZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | GRMZM2G139963_P01 |



