 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GRMZM2G169654_P01 | ||||||||
| Common Name | Zm.84705 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Andropogonodae; Andropogoneae; Tripsacinae; Zea
|
||||||||
| Family | RAV | ||||||||
| Protein Properties | Length: 389aa MW: 40493.6 Da PI: 9.9227 | ||||||||
| Description | RAV family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 36.9 | 9.4e-12 | 76 | 124 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
s+ykGV + +grW A+I++ r +r++lg+f + +Aa+a++ a+++++g
GRMZM2G169654_P01 76 SRYKGVVPQP-NGRWGAQIYE------RhQRVWLGTFAGEADAARAYDVAAQRFRG 124
78****9888.8*********......44**********99*************98 PP
| |||||||
| 2 | B3 | 101.1 | 6.3e-32 | 213 | 311 | 1 | 91 |
EEEE-..-HHHHTT-EE--HHH.HTT.......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE- CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh.......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkld 85
f+k+ tpsdv+k++rlv+pk++ae+h g + +++ l led g++W+++++y+++s++yvltkGW++Fvk++gL++gD+v F+++
GRMZM2G169654_P01 213 FDKTVTPSDVGKLNRLVIPKQHAEKHfplqlpsAGGESKGVLLNLEDAAGKVWRFRYSYWNSSQSYVLTKGWSRFVKEKGLQAGDVVGFYRS 304
89*******************************5556799**************************************************77 PP
SS.SEE. CS
B3 86 gr.sefe 91
+++
GRMZM2G169654_P01 305 AAgADTK 311
5545554 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF54171 | 4.77E-16 | 76 | 132 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 9.6E-8 | 76 | 124 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 1.18E-21 | 76 | 132 | No hit | No description |
| Gene3D | G3DSA:3.30.730.10 | 3.8E-19 | 77 | 132 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 1.4E-24 | 77 | 138 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 19.651 | 77 | 132 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:2.40.330.10 | 7.9E-40 | 208 | 322 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 1.57E-31 | 211 | 314 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 1.88E-30 | 211 | 314 | No hit | No description |
| SMART | SM01019 | 8.4E-24 | 213 | 319 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 8.0E-29 | 213 | 314 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 14.099 | 213 | 318 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 389 aa Download sequence Send to blast |
MDSASSLVDD TSSGGGGGAS TDKLRALAVF AAASGTPLER MGSGASAVVD AAEPGAEADS 60 GSGAAAVSVG GKLPSSRYKG VVPQPNGRWG AQIYERHQRV WLGTFAGEAD AARAYDVAAQ 120 RFRGRDAVTN FRPLADADPD AAAELRFLAS RSKAEVVDML RKHTYFDELA QNKRAFAAAS 180 ASAATASSLA NNPSSYASLS PATATAAARE HLFDKTVTPS DVGKLNRLVI PKQHAEKHFP 240 LQLPSAGGES KGVLLNLEDA AGKVWRFRYS YWNSSQSYVL TKGWSRFVKE KGLQAGDVVG 300 FYRSAAGADT KLFIDCKLRP NSVVVASTAG PSPPAPVAKA VRLFGVDLLT APATAAAPAE 360 AVAGCKRARD LGSPPQAAFK KQLVELALV |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wid_A | 5e-51 | 210 | 332 | 11 | 128 | DNA-binding protein RAV1 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Zm.84705 | 0.0 | cell culture| ear| endosperm| meristem| ovary| shoot | ||||
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | |||||
| Expression Atlas | GRMZM2G169654 | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00025 | PBM | Transfer from AT1G68840 | Download |
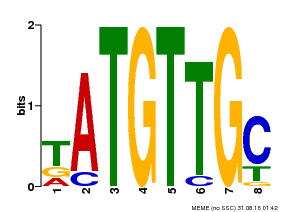
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | GRMZM2G169654_P01 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KJ727870 | 0.0 | KJ727870.1 Zea mays clone pUT5962 AP2-EREBP transcription factor (EREB126) gene, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001151105.2 | 0.0 | DNA-binding protein RAV1 | ||||
| Swissprot | Q8RZX9 | 0.0 | Y1934_ORYSJ; AP2/ERF and B3 domain-containing protein Os01g0693400 | ||||
| TrEMBL | A0A1D6NEV8 | 0.0 | A0A1D6NEV8_MAIZE; DNA-binding protein RAV1 | ||||
| STRING | GRMZM2G169654_P01 | 0.0 | (Zea mays) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP456 | 38 | 203 | Representative plant | OGRP365 | 16 | 108 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G68840.2 | 3e-97 | related to ABI3/VP1 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | GRMZM2G169654_P01 |



