 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSBRNA2T00007983001 | ||||||||
| Common Name | GSBRNA2T00007983001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 307aa MW: 33782.1 Da PI: 9.4417 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 38.5 | 2.6e-12 | 43 | 87 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r +WT+ E++++++a +++ + Wk+I + +g +t q++s+ qky
GSBRNA2T00007983001 43 RESWTEPEHDKFLEALQLFDRD-WKKIEAFIG-SKTVIQIRSHAQKY 87
789*****************77.*********.*************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 5.98E-16 | 37 | 93 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 13.876 | 38 | 92 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.8E-6 | 40 | 93 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 2.2E-18 | 41 | 90 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 9.6E-10 | 42 | 90 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 5.8E-10 | 43 | 87 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.21E-7 | 45 | 88 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 307 aa Download sequence Send to blast |
MNPADGSLLD PNGTMTVPAI GPLTSSEDLS KKIRKPYTIS KSRESWTEPE HDKFLEALQL 60 FDRDWKKIEA FIGSKTVIQI RSHAQKYFLK VQKSGTAEHL PPPRPKRKAA HPYPQKAQKN 120 VQPQVPGSFK STAEPNDPSY MFRPESSSML MTSPPPTAAA AAPWTNNVQT ISFTPLPKET 180 AGAGANNNCS SSSENTPRPR SNKDTNVQAN PGHSLRVLPD FGQVYSFIGS VFDPYASNHL 240 QKLKKMDPID VETVLLLMRN LSINLSSPDF EDHRRLLSSY DIGSETAADH GGVFKTLNKE 300 PPEIST* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Bna.27160 | 1e-103 | root| seed | ||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00560 | DAP | Transfer from AT5G52660 | Download |
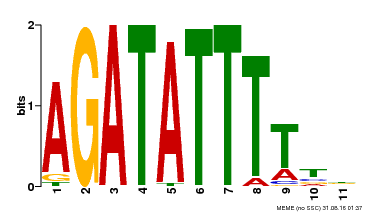
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | GSBRNA2T00007983001 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY550303 | 0.0 | AY550303.1 Arabidopsis thaliana MYB transcription factor (At5g52660) mRNA, complete cds. | |||
| GenBank | BT002002 | 0.0 | BT002002.1 Arabidopsis thaliana putative protein (At5g52660) mRNA, complete cds. | |||
| GenBank | BT025882 | 0.0 | BT025882.1 Arabidopsis thaliana At5g52660 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009119808.1 | 0.0 | PREDICTED: protein REVEILLE 6 isoform X1 | ||||
| Refseq | XP_013699049.1 | 0.0 | protein REVEILLE 6-like isoform X1 | ||||
| Swissprot | Q8H0W3 | 0.0 | RVE6_ARATH; Protein REVEILLE 6 | ||||
| TrEMBL | A0A078IS80 | 0.0 | A0A078IS80_BRANA; BnaA10g28420D protein | ||||
| TrEMBL | A0A397XKH6 | 0.0 | A0A397XKH6_BRACM; Uncharacterized protein | ||||
| STRING | Bra003117.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1290 | 28 | 93 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G52660.2 | 1e-153 | MYB_related family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



