 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSBRNA2T00011481001 | ||||||||
| Common Name | GSBRNA2T00011481001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 319aa MW: 35761 Da PI: 7.3662 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 170.1 | 7e-53 | 19 | 147 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpkkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdk 90
lppGfrFhPtdeelv +yLk+k+++++l++ +i++vd+yk++Pw+Lp+k+ +e+ewyfFs+rd+ky++g r+nra++sgyWkatg+dk
GSBRNA2T00011481001 19 LPPGFRFHPTDEELVIHYLKRKADSSPLPV-AIIADVDLYKHDPWELPEKALFGEQEWYFFSPRDRKYPNGARPNRAATSGYWKATGTDK 107
79****************************.88***************8888899*********************************** PP
NAM 91 evlsk..kgelvglkktLvfykgrapkgektdWvmheyrl 128
+v+s+ ++++vg+kk Lvfy+g+ pkg k+dW+mheyrl
GSBRNA2T00011481001 108 PVISTgdGNKKVGVKKALVFYTGKPPKGIKSDWIMHEYRL 147
****9888899***************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 7.59E-65 | 15 | 179 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 61.763 | 19 | 178 | IPR003441 | NAC domain |
| Pfam | PF02365 | 6.7E-28 | 20 | 147 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0045995 | Biological Process | regulation of embryonic development | ||||
| GO:0048317 | Biological Process | seed morphogenesis | ||||
| GO:0080060 | Biological Process | integument development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 319 aa Download sequence Send to blast |
MQVMETTDSS GGPPPQPNLP PGFRFHPTDE ELVIHYLKRK ADSSPLPVAI IADVDLYKHD 60 PWELPEKALF GEQEWYFFSP RDRKYPNGAR PNRAATSGYW KATGTDKPVI STGDGNKKVG 120 VKKALVFYTG KPPKGIKSDW IMHEYRLADN KPSLRCDFGH KKNSLRLDDW VLCRIYKKNN 180 STSSRHHHHH HLDNDKDHHH HFHHDMMRDD DLYRLPLMNI FPAVFSDNND PAAIYDGGGG 240 GAGYSMNHDF ASSSGLNQKP NIPMPFWDQD PAKRFNGGVG DCSDMVSSVA APSSMQQQGG 300 VLGDGLDRTS YHLTGLNW* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 3ulx_A | 2e-73 | 18 | 181 | 14 | 171 | Stress-induced transcription factor NAC1 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Bna.15799 | 0.0 | seed | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Expressed throughout anthers from stages 8 to 12. Later confined to distal region of anthers from stage 13. {ECO:0000269|PubMed:16055634}. | |||||
| Uniprot | TISSUE SPECIFICITY: Restricted primarily to the region of the embryo including the SAM (PubMed:12175016). Expressed in the outer integument, but seems not expressed in the embryo at the torpedo stage (PubMed:18849494). {ECO:0000269|PubMed:12175016, ECO:0000269|PubMed:18849494}. | |||||
| Uniprot | TISSUE SPECIFICITY: Stamen specific, in anthers from stage 8 (PubMed:15100403, PubMed:16055634). Expressed in the outer integument, but seems not expressed in the embryo at the torpedo stage (PubMed:18849494). {ECO:0000269|PubMed:15100403, ECO:0000269|PubMed:16055634, ECO:0000269|PubMed:18849494}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | May encode a transcription factor involved in the elaboration of shoot apical meristems (SAM) (Probable). Together with NAC056/NARS1, regulates embryogenesis by regulating the development and degeneration of ovule integuments, a process required for intertissue communication between the embryo and the maternal integument (PubMed:18849494). {ECO:0000269|PubMed:18849494, ECO:0000305}. | |||||
| UniProt | Transcription factor of the NAC family (Probable). Together with NAC018/NARS2, regulates embryogenesis by regulating the development and degeneration of ovule integuments, a process required for intertissue communication between the embryo and the maternal integument (PubMed:18849494). {ECO:0000269|PubMed:18849494, ECO:0000305}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00201 | DAP | Transfer from AT1G52880 | Download |
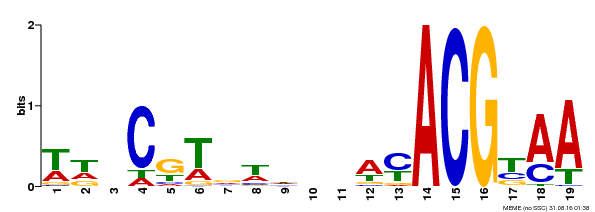
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | GSBRNA2T00011481001 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By Jasmonic acid (JA). {ECO:0000269|PubMed:16805732}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KF974757 | 0.0 | KF974757.1 Brassica napus NAC transcription factor 18-1 (NAC18-1.1) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009147651.1 | 0.0 | PREDICTED: NAC domain-containing protein 18 isoform X2 | ||||
| Swissprot | Q9LD44 | 1e-122 | NAC56_ARATH; NAC transcription factor 56 | ||||
| Swissprot | Q9ZNU2 | 1e-123 | NAC18_ARATH; NAC domain-containing protein 18 | ||||
| TrEMBL | A0A078IV55 | 0.0 | A0A078IV55_BRANA; BnaA06g01770D protein | ||||
| STRING | Bra018997.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM190 | 28 | 276 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G52880.1 | 1e-113 | NAC family protein | ||||



