 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSBRNA2T00020213001 | ||||||||
| Common Name | GSBRNA2T00020213001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 492aa MW: 54792.7 Da PI: 5.5476 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 42.8 | 9.2e-14 | 334 | 380 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCC.C...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPk.askapskKlsKaeiLekAveYIksLq 55
+h e+Er+RR+++N++f Lr ++P+ + K++Ka+ L A+ YIk+Lq
GSBRNA2T00020213001 334 NHVEAERQRREKLNQRFYALRAVVPNiS------KMDKASLLGDAISYIKELQ 380
799***********************66......******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 5.7E-51 | 50 | 231 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| PROSITE profile | PS50888 | 17.536 | 330 | 379 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.44E-18 | 333 | 393 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 1.73E-15 | 333 | 384 | No hit | No description |
| Pfam | PF00010 | 2.8E-11 | 334 | 380 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 1.2E-17 | 334 | 392 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.8E-17 | 336 | 385 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 492 aa Download sequence Send to blast |
MNTSDVVGWE DEDTRSMASA VLGNSASDFL TANSNSNQNL FLVMGTDDGL NEKLSRLVDS 60 PNPESFSWNY AVFWQQTVSR SGQQVLAWGD GCCREPKEEE SMAYNLEEEM RWQYMRKRVL 120 QKLHRMFGGS DEDDYALNLE NVTATEMFFL ASMYFFFNHG EGGPGRCFAS GRHVWLSDAV 180 GSDYCFRSFM VKSAGIRTVV MVPTDAGVLE LGSVWSLPEN VELVRSVQGL FARRVKPPNM 240 SGGKIHKLFG QELNSSHHNN NGSTIGYTSQ EIDVKVQENV NVVVVDDKNH KVMKTSCNEK 300 RPASLLPEGV SVVEEKRPRK RGRKPANGRE EALNHVEAER QRREKLNQRF YALRAVVPNI 360 SKMDKASLLG DAISYIKELQ ERVKIIEAEA SQEVVDVQAG EEEVVVRVVS PLESHPASRI 420 IQAMRNSEVV SVMESKLSLA EETLFHTFVV KNNNGSDPLT KEKVIAAAYP QTGLTQQLLL 480 PSSSSQVSGD I* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5gnj_A | 8e-27 | 328 | 392 | 2 | 66 | Transcription factor MYC2 |
| 5gnj_B | 8e-27 | 328 | 392 | 2 | 66 | Transcription factor MYC2 |
| 5gnj_E | 8e-27 | 328 | 392 | 2 | 66 | Transcription factor MYC2 |
| 5gnj_F | 8e-27 | 328 | 392 | 2 | 66 | Transcription factor MYC2 |
| 5gnj_G | 8e-27 | 328 | 392 | 2 | 66 | Transcription factor MYC2 |
| 5gnj_I | 8e-27 | 328 | 392 | 2 | 66 | Transcription factor MYC2 |
| 5gnj_M | 8e-27 | 328 | 392 | 2 | 66 | Transcription factor MYC2 |
| 5gnj_N | 8e-27 | 328 | 392 | 2 | 66 | Transcription factor MYC2 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 315 | 323 | KRPRKRGRK |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Bna.29213 | 1e-147 | seed | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: In seedlings, mainly expressed in root tips, cotyledons, hypocotyls, and leaves, as well as its guard cells. In adult plant, detectable in stems, flowers, and siliques. {ECO:0000269|PubMed:17828375}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator. Regulates positively abscisic acid (ABA) response. Confers drought tolerance and sensitivity to ABA. {ECO:0000269|PubMed:17828375}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00101 | PBM | Transfer from AT2G46510 | Download |
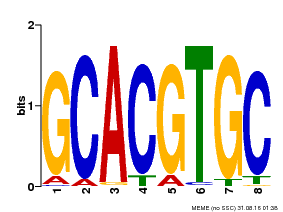
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | GSBRNA2T00020213001 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Transiently by ABA and polyethylene glycol (PEG). {ECO:0000269|PubMed:17828375}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013637084.1 | 0.0 | PREDICTED: transcription factor ABA-INDUCIBLE bHLH-TYPE-like isoform X1 | ||||
| Swissprot | Q9ZPY8 | 0.0 | AIB_ARATH; Transcription factor ABA-INDUCIBLE bHLH-TYPE | ||||
| TrEMBL | A0A078J3A8 | 0.0 | A0A078J3A8_BRANA; BnaC04g52120D protein | ||||
| STRING | Bo4g008180.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3758 | 27 | 59 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G46510.1 | 0.0 | ABA-inducible BHLH-type transcription factor | ||||



