 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSBRNA2T00031391001 | ||||||||
| Common Name | GSBRNA2T00031391001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | ARR-B | ||||||||
| Protein Properties | Length: 602aa MW: 66102.1 Da PI: 7.0787 | ||||||||
| Description | ARR-B family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 91 | 1e-28 | 202 | 255 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
kpr++W+ eLH++Fv av++L G +kA+Pk+ile+m+v+gLt+e+v+SHLQkYR+
GSBRNA2T00031391001 202 KPRVVWSVELHQQFVAAVNHL-GVDKAVPKKILEMMNVQGLTRENVASHLQKYRI 255
79*******************.********************************7 PP
| |||||||
| 2 | Response_reg | 74.2 | 5.1e-25 | 22 | 130 | 1 | 109 |
EEEESSSHHHHHHHHHHHHHTTCEEEEEESSHHHHHHHHHHHH..ESEEEEESSCTTSEHHHHHHHHHHHTTTSEEEEEESTTTHHHHHH CS
Response_reg 1 vlivdDeplvrellrqalekegyeevaeaddgeealellkekd..pDlillDiempgmdGlellkeireeepklpiivvtahgeeedale 88
vl+vdD+p+ +++l+++l+ y +v+ + + al+ll++++ +D+++ D+ mp+m+G++ll++ e +lp+i+++a ++++ +l+
GSBRNA2T00031391001 22 VLVVDDDPTCLMILERMLRTCLY-RVTKCNRAGIALTLLRKNKngFDIVISDVHMPDMNGFKLLEHVGLEM-DLPVIMMSADDSKSVVLK 109
89*********************.***************999889**********************6644.8***************** PP
HHHTTESEEEESS--HHHHHH CS
Response_reg 89 alkaGakdflsKpfdpeelvk 109
+ Ga d+l Kp+ +e+l +
GSBRNA2T00031391001 110 GVTHGAVDYLIKPVRIEALKN 130
*****************9986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PIRSF | PIRSF036392 | 5.2E-173 | 5 | 593 | IPR017053 | Response regulator B-type, plant |
| Gene3D | G3DSA:3.40.50.2300 | 7.1E-40 | 19 | 161 | No hit | No description |
| SuperFamily | SSF52172 | 4.84E-34 | 19 | 146 | IPR011006 | CheY-like superfamily |
| SMART | SM00448 | 2.1E-28 | 20 | 132 | IPR001789 | Signal transduction response regulator, receiver domain |
| PROSITE profile | PS50110 | 40.032 | 21 | 136 | IPR001789 | Signal transduction response regulator, receiver domain |
| Pfam | PF00072 | 6.5E-22 | 22 | 130 | IPR001789 | Signal transduction response regulator, receiver domain |
| CDD | cd00156 | 2.39E-26 | 23 | 135 | No hit | No description |
| PROSITE profile | PS51294 | 12.263 | 199 | 258 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.18E-19 | 200 | 259 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.8E-29 | 201 | 260 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 9.8E-25 | 202 | 255 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 2.1E-8 | 204 | 254 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009873 | Biological Process | ethylene-activated signaling pathway | ||||
| GO:0010082 | Biological Process | regulation of root meristem growth | ||||
| GO:0010119 | Biological Process | regulation of stomatal movement | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0071368 | Biological Process | cellular response to cytokinin stimulus | ||||
| GO:0080113 | Biological Process | regulation of seed growth | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 602 aa Download sequence Send to blast |
MLNPGVVGGS SNSDPFPAGL RVLVVDDDPT CLMILERMLR TCLYRVTKCN RAGIALTLLR 60 KNKNGFDIVI SDVHMPDMNG FKLLEHVGLE MDLPVIMMSA DDSKSVVLKG VTHGAVDYLI 120 KPVRIEALKN IWQHVVRKKR NVSEHSGSVE ETGGDRQQQQ RDDDDDGGGD NNSSSGNNEG 180 NLRKRKEEEQ GDDKEDTSSL KKPRVVWSVE LHQQFVAAVN HLGVDKAVPK KILEMMNVQG 240 LTRENVASHL QKYRIYLKRL GGVSQHQGNM NHSFMTGQDP SYGQLNGFDL QGLATAGQLP 300 AQSLAQLQAA GLGRSSSSLI KPGTMSVDQR SIFTFQNSKS RFGDGHGQMM MNGGSGNKQT 360 SLLHGVPTGH MRLQQQQMAG MRVADPSMQQ QQSMLSRRSL LDDAVVVRNS SRVLPGATHS 420 VFNNSFPLAS PPGMSVTDTK GGSSSATAAF CNPSYDILNN FPQQQHHNNN RVNEWDLPNM 480 GMVFNSHQDT TTAAFSASEA YSSSSTHKRK REAELVVEHG QNQQQPQSRS VNPMNQVYMN 540 GGGSVRVKTE TVTCPPQATT MFHEQYSNQE DLLSALLKQE GFPLLDSEFD FEGYSFDNIP 600 V* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 2e-23 | 198 | 260 | 1 | 63 | ARR10-B |
| Search in ModeBase | ||||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Detected in the whole plant. Predominantly expressed in pollen. {ECO:0000269|PubMed:11370868, ECO:0000269|PubMed:15173562, ECO:0000269|PubMed:9891419}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that binds specifically to the DNA sequence 5'-[AG]GATT-3'. Functions as a response regulator involved in His-to-Asp phosphorelay signal transduction system. Phosphorylation of the Asp residue in the receiver domain activates the ability of the protein to promote the transcription of target genes. Could directly activate some type-A response regulators in response to cytokinins. Involved in the expression of nuclear genes for components of mitochondrial complex I. Promotes cytokinin-mediated leaf longevity. Involved in the ethylene signaling pathway in an ETR1-dependent manner and in the cytokinin signaling pathway. {ECO:0000269|PubMed:11370868, ECO:0000269|PubMed:11574878, ECO:0000269|PubMed:15282545, ECO:0000269|PubMed:16407152}. | |||||
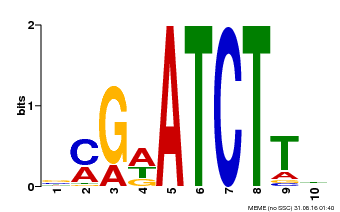
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00054 | PBM | Transfer from AT4G16110 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | GSBRNA2T00031391001 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC189632 | 0.0 | AC189632.2 Brassica rapa subsp. pekinensis cultivar Inbred line 'Chiifu' clone KBrS003O10, complete sequence. | |||
| GenBank | AC232469 | 0.0 | AC232469.1 Brassica rapa subsp. pekinensis cultivar Inbred line 'Chiifu' clone KBrB026A07, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013707663.1 | 0.0 | two-component response regulator ARR2-like isoform X2 | ||||
| Swissprot | Q9ZWJ9 | 0.0 | ARR2_ARATH; Two-component response regulator ARR2 | ||||
| TrEMBL | A0A078FCF1 | 0.0 | A0A078FCF1_BRANA; Two-component response regulator | ||||
| STRING | Bo7g104190.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2403 | 27 | 73 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16110.1 | 0.0 | response regulator 2 | ||||



