 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSBRNA2T00047942001 | ||||||||
| Common Name | GSBRNA2T00047942001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | B3 | ||||||||
| Protein Properties | Length: 487aa MW: 54867.3 Da PI: 5.8592 | ||||||||
| Description | B3 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 41.3 | 2.8e-13 | 23 | 91 | 17 | 98 |
E--HHH.HTT---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE..EEEEE- CS
B3 17 vlpkkfaeehggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrsefelvvkvfr 98
++ +f+++h ++ l + d+s + W+vkl ++++ + ++GW+eF+ka+ ++gD +vF++dg++ f +v+v
GSBRNA2T00047942001 23 TVDDEFLRKHTKV------LLISDSSDKIWKVKL--DGSR---LAGGGWEEFAKAHSFRDGDLLVFRHDGDEIF--HVSVSP 91
5566777777644......78889**********..8888...889**********************999999..777654 PP
| |||||||
| 2 | B3 | 67.4 | 2e-21 | 159 | 251 | 5 | 98 |
-..-HHHHTT-EE--HHH.HTT---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE..EE CS
B3 5 ltpsdvlksgrlvlpkkfaeehggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrsefelvv 94
+++++l ++rl l+k+f+ ++++++ ++ l +e+gr+W+++l+ + +sg +++++GW +F++angL +gD++ Fkl++ +++++v+
GSBRNA2T00047942001 159 RVTPYSLIKDRLDLSKDFTVVSFNEHNKPCEIDLVNEKGRKWTLRLSKNSTSGVFYIRQGWVNFCSANGLSQGDICKFKLSE-NGERPVL 247
578999999*********7666444556669***************77777777*************************987.6777999 PP
EEE- CS
B3 95 kvfr 98
+++
GSBRNA2T00047942001 248 RLCP 251
9986 PP
| |||||||
| 3 | B3 | 69.1 | 6.2e-22 | 279 | 376 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT---..--SEEEEEETTS-EEEEEE..EEETTE..EEE-TTHHHHHHHHT--TT-EEEEEE-SSS CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeehggkkeesktltledesgrsWevkliyrkksgr..yvltkGWkeFvkangLkegDfvvFkldgrs 88
f++v ++++ l++g+l+++ f++e g+kk s ++tl++++gr+W +l+++++ g+ ++l+ GW+e++kang+k++D++v++l++ +
GSBRNA2T00047942001 279 FLTVKLTPNRLQTGQLYISSVFVNESGIKK--SGEITLMNQDGRKWPSYLQMTGQCGSewFYLRHGWREMCKANGVKVNDSFVLELIW-E 365
77888999***************9999885..458***************9966655556****************************.7 PP
EE..EEEEE-S CS
B3 89 efelvvkvfrk 99
+ ++v+k+++k
GSBRNA2T00047942001 366 DDNPVFKFCSK 376
7789****987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50863 | 12.731 | 1 | 93 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 1.8E-12 | 10 | 93 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 3.2E-8 | 27 | 89 | IPR003340 | B3 DNA binding domain |
| SuperFamily | SSF101936 | 4.71E-12 | 27 | 91 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 1.0E-11 | 28 | 89 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 4.26E-10 | 28 | 89 | No hit | No description |
| SMART | SM01019 | 2.9E-24 | 155 | 253 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 14.819 | 155 | 252 | IPR003340 | B3 DNA binding domain |
| SuperFamily | SSF101936 | 3.73E-19 | 158 | 253 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 1.4E-18 | 160 | 252 | IPR015300 | DNA-binding pseudobarrel domain |
| Pfam | PF02362 | 3.0E-19 | 160 | 251 | IPR003340 | B3 DNA binding domain |
| CDD | cd10017 | 1.89E-19 | 165 | 251 | No hit | No description |
| Gene3D | G3DSA:2.40.330.10 | 2.6E-20 | 274 | 376 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 4.51E-20 | 278 | 378 | IPR015300 | DNA-binding pseudobarrel domain |
| SMART | SM01019 | 6.7E-28 | 279 | 377 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 5.0E-23 | 279 | 376 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 13.14 | 279 | 379 | IPR003340 | B3 DNA binding domain |
| CDD | cd10017 | 6.50E-16 | 279 | 375 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009908 | Biological Process | flower development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 487 aa Download sequence Send to blast |
MTIPPTFSLF HHRFITGDKP VLTVDDEFLR KHTKVLLISD SSDKIWKVKL DGSRLAGGGW 60 EEFAKAHSFR DGDLLVFRHD GDEIFHVSVS PRSDSCDISH HASPSLVDTD DVETDDDYSE 120 SDDGEDEYDD DDDGGEDDAG NIMVNKNTKA GFSCILRARV TPYSLIKDRL DLSKDFTVVS 180 FNEHNKPCEI DLVNEKGRKW TLRLSKNSTS GVFYIRQGWV NFCSANGLSQ GDICKFKLSE 240 NGERPVLRLC PSSNSHEEEE ECLEADARKG KKKNTPSQFL TVKLTPNRLQ TGQLYISSVF 300 VNESGIKKSG EITLMNQDGR KWPSYLQMTG QCGSEWFYLR HGWREMCKAN GVKVNDSFVL 360 ELIWEDDNPV FKFCSKAENK GNGNGRLRKK RACETSIVET ERRKRGRPRV SNTNSSNLQR 420 TRQESCSVSD QVASVKLSIL DTLNTVRQFR ADIEAREKNL EASSLEVDAL AERILGISQI 480 LNNNPV* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 388 | 404 | KKRACETSIVETERRKR |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Specifically expressed in the reproductive meristem. {ECO:0000269|PubMed:10092172}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | May play a role in flower development. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00602 | PBM | Transfer from AT4G31610 | Download |
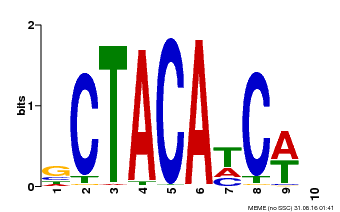
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | GSBRNA2T00047942001 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009108916.1 | 0.0 | PREDICTED: B3 domain-containing protein REM1-like isoform X2 | ||||
| Refseq | XP_013720407.1 | 0.0 | B3 domain-containing protein REM1-like isoform X2 | ||||
| Refseq | XP_022545721.1 | 0.0 | B3 domain-containing protein REM1-like isoform X2 | ||||
| Swissprot | O65098 | 0.0 | REM1_BRAOB; B3 domain-containing protein REM1 | ||||
| TrEMBL | A0A078JDF2 | 0.0 | A0A078JDF2_BRANA; BnaA08g30700D protein | ||||
| STRING | Bra010228.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM9950 | 17 | 36 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G31610.1 | 1e-173 | B3 family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



