 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSBRNA2T00047986001 | ||||||||
| Common Name | GSBRNA2T00047986001, LOC106417961, LOC106449689 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 301aa MW: 34515.2 Da PI: 7.8608 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 32.9 | 1.5e-10 | 14 | 61 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT.-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGgg.tWktIartmgkgRtlkqcksrwqky 47
+g+W++eEd +l+ +++ G+g +W + ++++g++R++k+c +++ +
GSBRNA2T00047986001 14 KGPWSPEEDAKLKSYIETSGTGgNWIALPHKIGLKRCGKSCMRWLNYL 61
79***************************************9777654 PP
| |||||||
| 2 | Myb_DNA-binding | 34 | 6.9e-11 | 69 | 110 | 3 | 46 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
+++ E+ ++ ++ G++ W+ Ia+ ++ gRt++++k++w++
GSBRNA2T00047986001 69 GFSEAEENIICNLYLTIGSR-WSIIAAQLP-GRTDNDIKNYWNT 110
699*****************.*********.***********97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 7.03E-22 | 11 | 108 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.0E-6 | 13 | 63 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.7E-9 | 14 | 61 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 4.5E-19 | 15 | 68 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.36E-5 | 16 | 61 | No hit | No description |
| PROSITE profile | PS51294 | 19.544 | 62 | 116 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.0E-9 | 66 | 114 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.1E-9 | 68 | 110 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.2E-22 | 69 | 117 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 2.45E-6 | 70 | 112 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 301 aa Download sequence Send to blast |
MGRSPCCDKA NVKKGPWSPE EDAKLKSYIE TSGTGGNWIA LPHKIGLKRC GKSCMRWLNY 60 LRPNIKHGGF SEAEENIICN LYLTIGSRWS IIAAQLPGRT DNDIKNYWNT RLKKKLIIKQ 120 RKELQEACIE QQEMMVMLKR QQQQIQPTFM MRQDQTMFTW PLQKLPFHHH NDGQVPTLVM 180 NSFGDQEDIK PEIIKNMVKI EDQEPERTNA FDHLYFSQLL LDPNSNYLGS GEGFSMNSIL 240 STNTNSPLRN TSISHQRFRN FHDEAVDLFL GEAGTSADQS TIRWEDISSL VYSDSKQVVN 300 * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 3e-20 | 14 | 116 | 7 | 108 | B-MYB |
| Search in ModeBase | ||||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Accumulates in an adaxial ball-shaped set of cells in three to five cell layers around the L3 layer of the shoot apical meristem (SAM) in youg plantlets. In the inflorescence meristem, confined to the axils of flower primordia. {ECO:0000269|PubMed:16461581}. | |||||
| Uniprot | TISSUE SPECIFICITY: Ubiquitous. {ECO:0000269|PubMed:16461581}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator (By similarity). Positively regulates axillary meristems (AMs) formation and development, especially during inflorescence. {ECO:0000250, ECO:0000269|PubMed:16461581}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00089 | SELEX | Transfer from AT3G49690 | Download |
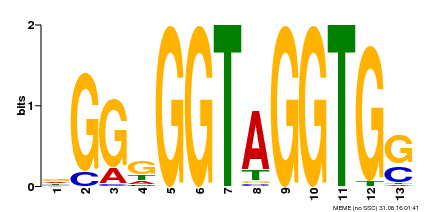
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | GSBRNA2T00047986001 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY519597 | 1e-165 | AY519597.1 Arabidopsis thaliana MYB transcription factor (At3g49690) mRNA, complete cds. | |||
| GenBank | BT029224 | 1e-165 | BT029224.1 Arabidopsis thaliana At3g49690 mRNA, complete cds. | |||
| GenBank | Y14209 | 1e-165 | Y14209.1 A.thaliana mRNA for AtMYB84 R2R3-MYB transcription factor. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013714118.1 | 0.0 | transcription factor RAX3-like | ||||
| Refseq | XP_013746869.1 | 0.0 | transcription factor RAX3-like | ||||
| Swissprot | Q9M2Y9 | 1e-161 | RAX3_ARATH; Transcription factor RAX3 | ||||
| TrEMBL | A0A078JGP4 | 0.0 | A0A078JGP4_BRANA; BnaC08g48600D protein | ||||
| TrEMBL | A0A3P6GJ21 | 0.0 | A0A3P6GJ21_BRAOL; Uncharacterized protein | ||||
| STRING | Bo8g078940.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2941 | 25 | 68 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G49690.1 | 1e-151 | myb domain protein 84 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 106417961 | 106449689 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



