 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSBRNA2T00050509001 | ||||||||
| Common Name | GSBRNA2T00050509001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 315aa MW: 35211.1 Da PI: 7.187 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 58.9 | 1.1e-18 | 14 | 61 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEde+lv +++ +G+g W+t ++ g++R++k+c++rw +yl
GSBRNA2T00050509001 14 KGPWTPEEDEKLVSYIQAHGPGKWRTLPKNAGLKRCGKSCRLRWTNYL 61
79********************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 52.8 | 9.4e-17 | 67 | 111 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
rg ++ +E+e +++++++lG++ W++Ia +++ gRt++++k++w+++
GSBRNA2T00050509001 67 RGEFSLQEEETIIQLHRLLGNK-WSAIAIHLP-GRTDNEIKNYWNTH 111
899*******************.*********.************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 3.2E-25 | 7 | 64 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 19.024 | 9 | 61 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.36E-30 | 12 | 108 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 5.2E-15 | 13 | 63 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.1E-16 | 14 | 61 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.28E-11 | 16 | 61 | No hit | No description |
| PROSITE profile | PS51294 | 25.84 | 62 | 116 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 7.2E-26 | 65 | 116 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 4.7E-16 | 66 | 114 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.6E-15 | 67 | 111 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.20E-11 | 69 | 112 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 315 aa Download sequence Send to blast |
MGKSSSSEES EVKKGPWTPE EDEKLVSYIQ AHGPGKWRTL PKNAGLKRCG KSCRLRWTNY 60 LRPDIKRGEF SLQEEETIIQ LHRLLGNKWS AIAIHLPGRT DNEIKNYWNT HIKKKLLRRG 120 IDPVTHCPRI NLLQLSSFLT SSLFKSMSQP VNIPFGLANT SINPELLNQL NTSLSNVQTE 180 SYQPHQANQQ LQNDHQTSFT GLLNSTPPVQ WQNNGECLEN YLNYTGSGDQ SINQAPLTGN 240 YSSAINDGEN YKVGWNFSSS MLPGTSSSSS TPLNSSSTAF INVGSEDDKE SYGSDMLMFH 300 HHHHDHDNNA LNLS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h8a_C | 3e-29 | 11 | 116 | 24 | 128 | MYB TRANSFORMING PROTEIN |
| Search in ModeBase | ||||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in rosette leaves, cauline leaves and flowers. {ECO:0000269|PubMed:8980549}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may function in osmotic stress and wounding signaling pathways (Probable). Contributes to basal resistance against the herbivore Pieris rapae (white cabbage butterfly) feeding (PubMed:19517001). {ECO:0000269|PubMed:19517001, ECO:0000305|PubMed:12857823}. | |||||
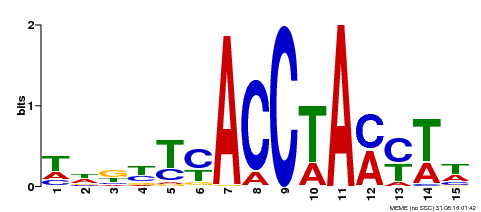
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00563 | DAP | Transfer from AT5G54230 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | GSBRNA2T00050509001 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by light (PubMed:8980549). Induced by wounding, salt stress and abscisic acid (PubMed:12857823). Induced by the lepidopteran herbivore Pieris rapae (white cabbage butterfly) feeding (PubMed:19517001). {ECO:0000269|PubMed:12857823, ECO:0000269|PubMed:19517001, ECO:0000269|PubMed:8980549}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009132444.1 | 0.0 | PREDICTED: transcription factor MYB3 | ||||
| Swissprot | Q9LDR8 | 7e-83 | MY102_ARATH; Transcription factor MYB102 | ||||
| TrEMBL | A0A078H4P4 | 0.0 | A0A078H4P4_BRANA; BnaC09g13310D protein | ||||
| STRING | Bra029033.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM14465 | 17 | 22 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G54230.1 | 1e-150 | myb domain protein 49 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



