 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSBRNA2T00051682001 | ||||||||
| Common Name | GSBRNA2T00051682001, LOC106446414 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 452aa MW: 50254.9 Da PI: 7.1279 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 38.9 | 1.5e-12 | 304 | 350 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCC.C...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPk.askapskKlsKaeiLekAveYIksLq 55
+h e+Er+RR+++N++f Lr ++P+ + K++Ka+ L A+ YI ++q
GSBRNA2T00051682001 304 NHVEAERQRREKLNQRFYALRAVVPNiS------KMDKASLLADAITYITDMQ 350
799***********************66......***************9998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 3.1E-40 | 47 | 216 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| PROSITE profile | PS50888 | 16.234 | 300 | 349 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.44E-17 | 300 | 366 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 3.02E-14 | 303 | 354 | No hit | No description |
| Pfam | PF00010 | 4.0E-10 | 304 | 350 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 5.6E-17 | 304 | 366 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.6E-14 | 306 | 355 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 452 aa Download sequence Send to blast |
MGQKFWDNQE DRAMVESTIG SEACDFFISS ASSLTKLLPP PPTDPNLQQG LRHVVEGSDW 60 DYAIFWLASN VNSSDGCVLI WGDGHCRVVK GNSGDEQDET KRRVLSKLHL SFVGSDLVKQ 120 GPLTDLDMFY LASLYFSFRC DSNKYGPAGT YVSGKPLWAA DLPSCLSYYR VRSFLARSAG 180 FKTVLSVPVN CGVVELGSLK LIPEDKSVVE MVKSVFGGSD FVKTKEAPKI FGRQLSLGGS 240 KPRSMSINFS PKVEDDSGFS LEAYEVGGSN QVYGKDEAAL YLTDEQRPRK RGRKPANGRE 300 EALNHVEAER QRREKLNQRF YALRAVVPNI SKMDKASLLA DAITYITDMQ KKIRVYETEK 360 QVMKRRESNQ ITPAEVDYQQ RQDDAVVRVS CPLENHPVSK VIQVFKENEV TPHDANVAVT 420 EEGVVHTFTL RPQGGCTAEQ LKDKLLTSLA Q* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5gnj_A | 5e-26 | 298 | 381 | 2 | 81 | Transcription factor MYC2 |
| 5gnj_B | 5e-26 | 298 | 381 | 2 | 81 | Transcription factor MYC2 |
| 5gnj_E | 5e-26 | 298 | 381 | 2 | 81 | Transcription factor MYC2 |
| 5gnj_F | 5e-26 | 298 | 381 | 2 | 81 | Transcription factor MYC2 |
| 5gnj_G | 5e-26 | 298 | 381 | 2 | 81 | Transcription factor MYC2 |
| 5gnj_I | 5e-26 | 298 | 381 | 2 | 81 | Transcription factor MYC2 |
| 5gnj_M | 5e-26 | 298 | 381 | 2 | 81 | Transcription factor MYC2 |
| 5gnj_N | 5e-26 | 298 | 381 | 2 | 81 | Transcription factor MYC2 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Bna.17215 | 0.0 | root| seed | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00099 | PBM | Transfer from AT4G16430 | Download |
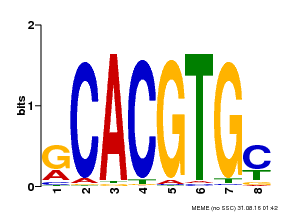
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | GSBRNA2T00051682001 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By UV treatment. {ECO:0000269|PubMed:12679534}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013743591.1 | 0.0 | transcription factor bHLH3 | ||||
| Swissprot | O23487 | 0.0 | BH003_ARATH; Transcription factor bHLH3 | ||||
| TrEMBL | A0A078H403 | 0.0 | A0A078H403_BRANA; BnaA01g17420D protein | ||||
| STRING | Bra038458.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM6611 | 27 | 41 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16430.1 | 0.0 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 106446414 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



