 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSBRNA2T00060177001 | ||||||||
| Common Name | GSBRNA2T00060177001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | GRAS | ||||||||
| Protein Properties | Length: 557aa MW: 60610.6 Da PI: 5.6662 | ||||||||
| Description | GRAS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GRAS | 445.3 | 4.1e-136 | 191 | 551 | 1 | 374 |
GRAS 1 lvelLlecAeavssgdlelaqalLarlselaspdgdpmqRlaayfteALaarlarsvselykalppset.seknsseelaalklfsevsP 89
lv++L++cAeav+s++l+la+al++++ la +++ +m+++a+yf+eALa+r++r l+p +t +++ s++l++ f+e++P
GSBRNA2T00060177001 191 LVHALMACAEAVQSSNLTLAEALVKQIGFLAVSQAGAMRKVATYFAEALARRIYR--------LSPPQTqIDHSLSDTLQM--HFYETCP 270
689****************************************************........444444043445555444..5****** PP
GRAS 90 ilkfshltaNqaIleavegeervHiiDfdisqGlQWpaLlqaLasRpegppslRiTgvgspesgskeeleetgerLakfAeelgvpfefn 179
+lkf+h+taNqaIlea+eg++rvH+iDf+++qGlQWpaL+qaLa R++gpps+R+Tg+g+p++++++ l+e+g +La++Ae+++v+fe++
GSBRNA2T00060177001 271 YLKFAHFTANQAILEAFEGKKRVHVIDFSMNQGLQWPALMQALALREGGPPSFRLTGIGPPAADNSDHLHEVGCKLAQLAEAIHVEFEYR 360
****************************************************************************************** PP
GRAS 180 vlvakrledleleeLrvkp..gEalaVnlvlqlhrlldesvsleserdevLklvkslsPkvvvvveqeadhnsesFlerflealeyysal 267
+va++l+dl+ ++L+++p E++aVn+v++lh+ll +++ +e+ v +vk+++P +++vveqe++hn++ Fl+rf+e+l+yys+l
GSBRNA2T00060177001 361 GFVANSLADLDASMLELRPseTESVAVNSVFELHKLLGRPGGIEK----VFGVVKQIKPVIFTVVEQESNHNGPVFLDRFTESLHYYSTL 446
******************99899******************9999....***************************************** PP
GRAS 268 fdsleaklpreseerikvErellgreivnvvacegaerrerhetlekWrerleeaGFkpvplsekaakqaklllrkvk.sdgyrveeesg 356
fdsle s++++++++++lg++i+n+vaceg +r+erhetl++W++r++++GF p++l+++a kqa++ll+ ++ ++gyrvee++g
GSBRNA2T00060177001 447 FDSLEGA---PSSQDKVMSEVYLGKQICNLVACEGPDRVERHETLSQWSNRFGSSGFAPAHLGSNAFKQASTLLALFNgGEGYRVEENNG 533
****999...79999*************************************************************************** PP
GRAS 357 slvlgWkdrpLvsvSaWr 374
+l+l W++rpL+++SaW+
GSBRNA2T00060177001 534 CLMLSWHTRPLITTSAWK 551
*****************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01129 | 1.3E-39 | 42 | 115 | No hit | No description |
| Pfam | PF12041 | 9.4E-33 | 42 | 109 | IPR021914 | Transcriptional factor DELLA, N-terminal |
| PROSITE profile | PS50985 | 66.001 | 165 | 530 | IPR005202 | Transcription factor GRAS |
| Pfam | PF03514 | 1.4E-133 | 191 | 551 | IPR005202 | Transcription factor GRAS |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009867 | Biological Process | jasmonic acid mediated signaling pathway | ||||
| GO:0009938 | Biological Process | negative regulation of gibberellic acid mediated signaling pathway | ||||
| GO:0010187 | Biological Process | negative regulation of seed germination | ||||
| GO:0010218 | Biological Process | response to far red light | ||||
| GO:0042176 | Biological Process | regulation of protein catabolic process | ||||
| GO:0042538 | Biological Process | hyperosmotic salinity response | ||||
| GO:2000033 | Biological Process | regulation of seed dormancy process | ||||
| GO:2000377 | Biological Process | regulation of reactive oxygen species metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 557 aa Download sequence Send to blast |
MKRDLHQFQG PNHGTSIAGS STSSPAVFGK DKMMMVKEEE DDELLGVLGY KVRSSEMAEV 60 ALKLEQLETM MGNVQEDGLA HLATDTVHYN PAELYSWLDN MLTELNPPAA SGSSGDYDLK 120 AIPGSAICRG SNQFAVDSLN NKRLKPCSSP DSMVTSPSPA GVIGTTVTTT TASESTRPLI 180 LVDSQDNGVR LVHALMACAE AVQSSNLTLA EALVKQIGFL AVSQAGAMRK VATYFAEALA 240 RRIYRLSPPQ TQIDHSLSDT LQMHFYETCP YLKFAHFTAN QAILEAFEGK KRVHVIDFSM 300 NQGLQWPALM QALALREGGP PSFRLTGIGP PAADNSDHLH EVGCKLAQLA EAIHVEFEYR 360 GFVANSLADL DASMLELRPS ETESVAVNSV FELHKLLGRP GGIEKVFGVV KQIKPVIFTV 420 VEQESNHNGP VFLDRFTESL HYYSTLFDSL EGAPSSQDKV MSEVYLGKQI CNLVACEGPD 480 RVERHETLSQ WSNRFGSSGF APAHLGSNAF KQASTLLALF NGGEGYRVEE NNGCLMLSWH 540 TRPLITTSAW KLSAVH* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5b3g_A | 6e-62 | 198 | 550 | 26 | 378 | Protein SCARECROW |
| 5b3h_A | 6e-62 | 198 | 550 | 25 | 377 | Protein SCARECROW |
| 5b3h_D | 6e-62 | 198 | 550 | 25 | 377 | Protein SCARECROW |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Bna.12264 | 0.0 | leaf| root| seed | ||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcriptional regulator that acts as a repressor of the gibberellin (GA) signaling pathway. Probably acts by participating in large multiprotein complexes that repress transcription of GA-inducible genes. Upon GA application, it is degraded by the proteasome, allowing the GA signaling pathway. {ECO:0000269|PubMed:15734906}. | |||||
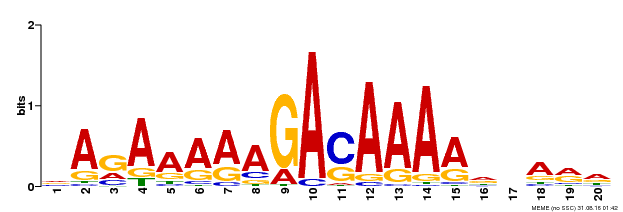
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00611 | ChIP-seq | Transfer from AT2G01570 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | GSBRNA2T00060177001 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY928549 | 0.0 | AY928549.1 Brassica rapa DELLA protein (RGA1) gene, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013598468.1 | 0.0 | PREDICTED: DELLA protein RGA1 | ||||
| Swissprot | Q5BN23 | 0.0 | RGA1_BRACM; DELLA protein RGA1 | ||||
| TrEMBL | A0A078HEN8 | 0.0 | A0A078HEN8_BRANA; BnaC07g20900D protein | ||||
| STRING | Bo7g078550.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM837 | 28 | 121 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G01570.1 | 0.0 | GRAS family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



