 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSBRNA2T00061848001 | ||||||||
| Common Name | GSBRNA2T00061848001, LOC106411337 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 546aa MW: 60897.6 Da PI: 5.2998 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 154.8 | 3.9e-48 | 45 | 171 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLp.k.kvkaeekewyfFskrdkkyatgkrknratksgyWkatgk 88
l+pGfrFhPtdeelvv+yLk+k+++kkl++ +i e+d+yk++P++Lp k +++++++w+fFs+rd+k+ +g r++rat++gyWkatg
GSBRNA2T00061848001 45 LAPGFRFHPTDEELVVYYLKRKIRRKKLRV-AAIGETDVYKFDPEELPgKaLHNTGDRQWFFFSPRDRKQ-HGGRSSRATDRGYWKATGV 132
579***************************.88***************6434556889**********97.699**************** PP
NAM 89 dkevlskkgelvglkktLvfykgrapkgektdWvmheyrl 128
d+ +++ +++ vg kktLvf++grapkge+tdWvmhey+l
GSBRNA2T00061848001 133 DRIIKC-NSRPVGEKKTLVFHRGRAPKGERTDWVMHEYTL 171
*****9.999****************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 1.57E-55 | 40 | 194 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 52.869 | 45 | 194 | IPR003441 | NAC domain |
| Pfam | PF02365 | 2.0E-26 | 47 | 171 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010114 | Biological Process | response to red light | ||||
| GO:0010224 | Biological Process | response to UV-B | ||||
| GO:0031930 | Biological Process | mitochondria-nucleus signaling pathway | ||||
| GO:1900409 | Biological Process | positive regulation of cellular response to oxidative stress | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005789 | Cellular Component | endoplasmic reticulum membrane | ||||
| GO:0016021 | Cellular Component | integral component of membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 546 aa Download sequence Send to blast |
MQSYWNIRSS SQILNLHQQK SLGFFFSSPV METSRGSCRA GDRVLAPGFR FHPTDEELVV 60 YYLKRKIRRK KLRVAAIGET DVYKFDPEEL PGKALHNTGD RQWFFFSPRD RKQHGGRSSR 120 ATDRGYWKAT GVDRIIKCNS RPVGEKKTLV FHRGRAPKGE RTDWVMHEYT LHKEELEKCG 180 DVKDNFVLYK IFKKSGSGPK NGEHYGAPFV EEEWAEEDDE VDEADAVHVP TNQLMVSVCL 240 GSNNNIWADG GLNQSELNEN DIQELMRQVS EETGVNSHVA NNNPVNLAED EYLEIDDLLL 300 PGPEPSYVDK EGSAVLNDND FFDVDSYIGD FDATNPQSVP VGVGLNNGVV QSLPVNTFPV 360 TDQANNNQFQ HQTWKNQDSN WPLRNSYTRK ISSGSWTPEL NNNEVTICKF GEAPGDDASE 420 FINPLTSAKG EEATKDESSQ FSSSVWSFLE SIPASPAFAS ENPIVNLNIV RISSLGGRYR 480 FGSKSTSTNV VIAVNDSEAE SKKSGGYNKK NNHKGFFCLS IIGALCALSW VMMGTMGVSG 540 RSLLW* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 3swm_A | 9e-44 | 45 | 200 | 20 | 174 | NAC domain-containing protein 19 |
| 3swm_B | 9e-44 | 45 | 200 | 20 | 174 | NAC domain-containing protein 19 |
| 3swm_C | 9e-44 | 45 | 200 | 20 | 174 | NAC domain-containing protein 19 |
| 3swm_D | 9e-44 | 45 | 200 | 20 | 174 | NAC domain-containing protein 19 |
| 3swp_A | 9e-44 | 45 | 200 | 20 | 174 | NAC domain-containing protein 19 |
| 3swp_B | 9e-44 | 45 | 200 | 20 | 174 | NAC domain-containing protein 19 |
| 3swp_C | 9e-44 | 45 | 200 | 20 | 174 | NAC domain-containing protein 19 |
| 3swp_D | 9e-44 | 45 | 200 | 20 | 174 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 65 | 72 | KIRRKKLR |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Bna.27232 | 0.0 | seed | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in roots, rosette leaves, shoot apex, stems and flowers. {ECO:0000269|PubMed:17158162}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator activated by proteolytic cleavage through regulated intramembrane proteolysis (RIP). Involved in oxidative stress tolerance by mediating regulation of mitochondrial retrograde signaling during mitochondrial dysfunction. Interacts directly with the mitochondrial dysfunction DNA consensus motif 5'-CTTGNNNNNCA[AC]G-3', a cis-regulatory elements of several mitochondrial retrograde regulation-induced genes, and triggers increased oxidative stress tolerance. {ECO:0000269|PubMed:24045019}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00176 | DAP | Transfer from AT1G32870 | Download |
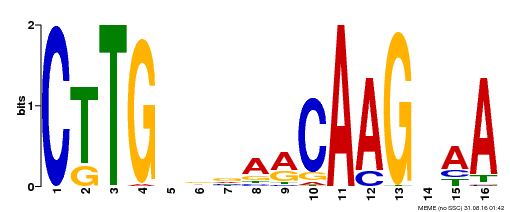
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | GSBRNA2T00061848001 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat, salt stress, abscisic acid (ABA) and methyl methanesulfonate (MMS) treatment. {ECO:0000269|PubMed:17158162}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KF974756 | 0.0 | KF974756.1 Brassica napus NAC transcription factor 13-1 (NAC13-1.1) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013707538.1 | 0.0 | NAC domain-containing protein 13 isoform X1 | ||||
| Refseq | XP_013707539.1 | 0.0 | NAC domain-containing protein 13 isoform X2 | ||||
| Refseq | XP_013707541.1 | 0.0 | NAC domain-containing protein 13 isoform X4 | ||||
| Swissprot | F4IED2 | 0.0 | NAC13_ARATH; NAC domain-containing protein 13 | ||||
| TrEMBL | A0A078FDS7 | 0.0 | A0A078FDS7_BRANA; BnaC07g13550D protein | ||||
| STRING | Bo7g057730.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM15317 | 15 | 17 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G32870.1 | 1e-172 | NAC domain protein 13 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 106411337 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



