 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSBRNA2T00068139001 | ||||||||
| Common Name | GSBRNA2T00068139001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 1326aa MW: 148033 Da PI: 7.2028 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 11.2 | 0.0011 | 1208 | 1233 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
y+C C++sFs+ +L H r+ +
GSBRNA2T00068139001 1208 YQCDmeGCTMSFSSEKQLSLHKRNiC 1233
99********************9877 PP
| |||||||
| 2 | zf-C2H2 | 16.5 | 2.4e-05 | 1233 | 1255 | 3 | 23 |
ET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 3 Cp..dCgksFsrksnLkrHirtH 23
Cp Cgk+F ++ +L++H+r+H
GSBRNA2T00068139001 1233 CPvkGCGKTFFSHKYLVQHQRVH 1255
9999*****************99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00545 | 4.1E-17 | 19 | 60 | IPR003349 | JmjN domain |
| PROSITE profile | PS51183 | 14.284 | 20 | 61 | IPR003349 | JmjN domain |
| Pfam | PF02375 | 1.6E-15 | 21 | 54 | IPR003349 | JmjN domain |
| SMART | SM00558 | 3.0E-53 | 200 | 369 | IPR003347 | JmjC domain |
| PROSITE profile | PS51184 | 33.783 | 203 | 369 | IPR003347 | JmjC domain |
| SuperFamily | SSF51197 | 1.02E-27 | 215 | 386 | No hit | No description |
| Pfam | PF02373 | 1.7E-37 | 233 | 352 | IPR003347 | JmjC domain |
| Gene3D | G3DSA:3.30.160.60 | 9.7E-4 | 1207 | 1230 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 9.4 | 1208 | 1230 | IPR015880 | Zinc finger, C2H2-like |
| SMART | SM00355 | 0.0015 | 1231 | 1255 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 13.339 | 1231 | 1260 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 6.9E-7 | 1232 | 1259 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 1233 | 1255 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 1.3E-10 | 1247 | 1289 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 9.0E-10 | 1260 | 1284 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 11.801 | 1261 | 1290 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0015 | 1261 | 1285 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 1263 | 1285 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 2.47E-8 | 1279 | 1313 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 1.2E-9 | 1285 | 1314 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 2.2 | 1291 | 1317 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.388 | 1291 | 1322 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1293 | 1317 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009826 | Biological Process | unidimensional cell growth | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0033169 | Biological Process | histone H3-K9 demethylation | ||||
| GO:0035067 | Biological Process | negative regulation of histone acetylation | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1326 aa Download sequence Send to blast |
MAVSEQSQDV FPWLKSLPVA PEFRPTLAEF QDPMAYICKI EEEASRYGIC KIVPPVPPSS 60 KKTAINNLNR SLAARARARA RDGNGGKSDY DGGPTFTTRQ QQIGFCPRRQ RPVQRPVWQS 120 GERYTFDEFE FKAKNFEKSY LKKCGKKGSV SPLEVETLYW RASVDKPFSV EYANDMPGSA 180 FVPLSLAAAR RRESGGDCGT VGETAWNMRA MARAEGSLLK FMKEEIPGVT SPMVYIAMMF 240 SWFAWHVEDH DLHSLNYLHM GAAKTWYGVP KDAAMAFEEV VRVHGYGEEL NPLVTFSTLG 300 EKTTVMSPEV FVRAGIPCCR LVQNPGDFVI TFPGAYHSGF SHGFNFGEAS NIATPQWLRM 360 AKDAAIRRAS INYPPMVSHL QLLYDYALAL GSRVPDSIHN KPRSSRLKDK KKSEGEKLTN 420 ELFVQNVIHN NELLHSLGKG SPIALLPQSS SDVSVCSDVR IGSHLGANQG QTTLLIKSED 480 LKKFTSLCER NRDHLASKED ETQGTSTDGE KRKNNGAVGL SDQRLFSCVT CGVLSFDCVA 540 IIQPKEAAAR YLMSADCSFL NDWTVASGSA NLGQDVVMPP SENTGKQDVG DLYNAPVQTP 600 YHSTTKTVDQ RTSSSSLTKE NGALGLLASA YGDSSDSEEE DHKGLDNPVS DEVEASSFVT 660 DGNDEAGNGL SSALNSQGLT CEKGKEVDVS HANLSKGGNT SSVEITLPFI PRSDDDFSRL 720 HVFCLEHAAE VEQRLHPIGG INIMLLCHPD YPRIEAEGKV VAKELGVNHE WNDTEFKNVT 780 HEETIQAALA NVEAKAGNSD WAVKLGINLS HSAILSRSPL YSKQMPYNSV IYNVFGRTSP 840 ATPQLSGIRS SRQRKYVAGK WCGKVWMSHQ VHPFLLEEDL EGEETERSHL RAALDEDVTV 900 HGNDSRDATT MFGRKYSRKR KARGKAAPRK KLTSFKREAG VSDDTTSEDH SYKQQWRAYG 960 DEEESYFETG NEVSGDSSNQ MSDQQQLKGV ESDDDEVSER SLGQEYAVRR EYASSESSME 1020 NGFQVYREDQ PMYGDDDMYR HPRGIPRNKR TKVFRDLVSY DSEDQQRERV FTSDAQTSRM 1080 GDEYDSEENS LEEQDFCSSG KRQTRSIAKR KVKTKIVQSL GDTGGHTLLQ SGSRKKMKEL 1140 DSYMEGPSTR LRVRTPKPSR GSSATKPKKT GKKGRNVSFS EVASEEEVEE NEEESEEAST 1200 RIRECLPYQC DMEGCTMSFS SEKQLSLHKR NICPVKGCGK TFFSHKYLVQ HQRVHSDDRP 1260 LKCPWKGCKM TFKWAWSRTE HIRVHTGERP YICAEPGCSQ TFRFVSDFSR HKRKTGHSAK 1320 KIKKK* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6a57_A | 3e-73 | 1205 | 1325 | 20 | 140 | Lysine-specific demethylase REF6 |
| 6a58_A | 3e-73 | 1205 | 1325 | 20 | 140 | Lysine-specific demethylase REF6 |
| 6a59_A | 3e-73 | 1205 | 1325 | 20 | 140 | Lysine-specific demethylase REF6 |
| 6ip0_A | 1e-70 | 9 | 392 | 4 | 355 | Transcription factor jumonji (Jmj) family protein |
| 6ip4_A | 1e-70 | 9 | 392 | 4 | 355 | Arabidopsis JMJ13 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 918 | 930 | KRKARGKAAPRKK |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Highly expressed in the shoot apical meristem and primary and secondary root tips, and lower expression in cotyledons, leaves and root axis along vascular tissues. Detected in inflorescences, stems and siliques. {ECO:0000269|PubMed:15377760, ECO:0000269|PubMed:18713399}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Histone demethylase that demethylates 'Lys-27' (H3K27me) of histone H3. Demethylates both tri- (H3K27me3) and di-methylated (H3K27me2) H3K27me (PubMed:21642989, PubMed:27111035). Demethylates also H3K4me3/2 and H3K36me3/2 in an in vitro assay (PubMed:20711170). Involved in the transcriptional regulation of hundreds of genes regulating developmental patterning and responses to various stimuli (PubMed:18467490). Binds DNA via its four zinc fingers in a sequence-specific manner, 5'-CTCTGYTY-3', to promote the demethylation of H3K27me3 and the regulation of organ boundary formation (PubMed:27111034, PubMed:27111035). Involved in the regulation of flowering time by repressing FLOWERING LOCUS C (FLC) expression (PubMed:15377760). Interacts with the NF-Y complexe to regulate SOC1 (PubMed:25105952). Mediates the recruitment of BRM to its target loci (PubMed:27111034). {ECO:0000269|PubMed:15377760, ECO:0000269|PubMed:18467490, ECO:0000269|PubMed:20711170, ECO:0000269|PubMed:21642989, ECO:0000269|PubMed:25105952, ECO:0000269|PubMed:27111034, ECO:0000269|PubMed:27111035}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00608 | ChIP-seq | Transfer from AT3G48430 | Download |
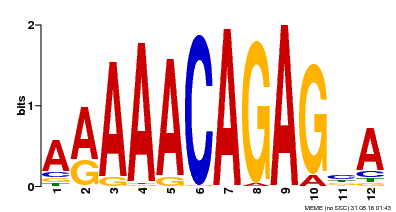
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | GSBRNA2T00068139001 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013724852.1 | 0.0 | lysine-specific demethylase REF6 | ||||
| Swissprot | Q9STM3 | 0.0 | REF6_ARATH; Lysine-specific demethylase REF6 | ||||
| TrEMBL | A0A078HQG9 | 0.0 | A0A078HQG9_BRANA; BnaCnng09530D protein | ||||
| STRING | Bo8g076910.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM5259 | 28 | 43 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48430.1 | 0.0 | relative of early flowering 6 | ||||



