 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSBRNA2T00069535001 | ||||||||
| Common Name | GSBRNA2T00069535001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 522aa MW: 58204.1 Da PI: 7.6973 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 57.9 | 2.3e-18 | 73 | 119 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g WT+eEde l +av ++ g++Wk+Ia++++ Rt+ qc +rwqk+l
GSBRNA2T00069535001 73 KGGWTPEEDETLRQAVDKYKGKRWKKIAEYFP-ERTEVQCLHRWQKVL 119
688*****************************.************986 PP
| |||||||
| 2 | Myb_DNA-binding | 58 | 2.1e-18 | 125 | 171 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g WT+eEd ++v++vk++G+ W++Ia+ ++ gR +kqc++rw+++l
GSBRNA2T00069535001 125 KGTWTQEEDGIIVELVKKYGPAKWSLIAKSLP-GRIGKQCRERWHNHL 171
799*****************************.*************97 PP
| |||||||
| 3 | Myb_DNA-binding | 55.6 | 1.2e-17 | 177 | 220 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
+++WTteE+ l++a + +G++ W+ Ia+ ++ gRt++ +k++w++
GSBRNA2T00069535001 177 KDAWTTEEETALINAQRTHGNK-WAEIAKVLP-GRTDNAIKNHWNS 220
679*******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 19.28 | 68 | 119 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 3.59E-16 | 69 | 126 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.9E-16 | 72 | 121 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 8.1E-17 | 73 | 119 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 6.6E-25 | 75 | 135 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 2.04E-14 | 76 | 119 | No hit | No description |
| PROSITE profile | PS51294 | 30.89 | 120 | 175 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 9.9E-31 | 122 | 218 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.9E-16 | 124 | 173 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 6.8E-17 | 125 | 171 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.45E-14 | 127 | 171 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 5.4E-26 | 136 | 178 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 21.801 | 176 | 226 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.8E-16 | 176 | 224 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.0E-15 | 177 | 220 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 9.8E-23 | 179 | 226 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.52E-12 | 179 | 222 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 522 aa Download sequence Send to blast |
MSSGSNPLAC SPEKEEEERS EMKIEILCME NKQPTPASSE GSASIFLYSP QLKTPAPASP 60 SHRRTSGPMR RAKGGWTPEE DETLRQAVDK YKGKRWKKIA EYFPERTEVQ CLHRWQKVLN 120 PELIKGTWTQ EEDGIIVELV KKYGPAKWSL IAKSLPGRIG KQCRERWHNH LNPGIRKDAW 180 TTEEETALIN AQRTHGNKWA EIAKVLPGRT DNAIKNHWNS SLKKKLDFYL ATCNLPPSAT 240 KIGDFADVDR DFKLSGATKP LKDSDSVTQS SSVNTDANED GVDHVHSSSD LLEESAVNEY 300 ACSPASVECK PQLPNLRPMS PRINSKDFVQ KKEENGLGTP RHGNLYYKSP LDFFYFPSEA 360 DLRRMYGYEC GCSPSAVSSS PVSLMITPPC NKDSGLAAAT RSPESFLREA ARTFPNTPSI 420 FRKRRKVVLA NKTDDEDDAV NSVAKGADQN ENSKDSAEIS TVGREALLLL ETVDDSNGST 480 FNASPPYRVR AKRRAVFKSR QLEFTSEKEN QPETSEEDKS V* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h88_C | 1e-66 | 72 | 226 | 5 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 1e-66 | 72 | 226 | 5 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| Search in ModeBase | ||||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Expressed both in proliferating and maturing stages of leaves. {ECO:0000269|PubMed:26069325}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds 5'-AACGG-3' motifs in gene promoters (By similarity). Transcription repressor that regulates organ growth. Binds to the promoters of G2/M-specific genes and to E2F target genes to prevent their expression in post-mitotic cells and to restrict the time window of their expression in proliferating cells (PubMed:26069325). {ECO:0000250|UniProtKB:Q94FL9, ECO:0000269|PubMed:26069325}. | |||||
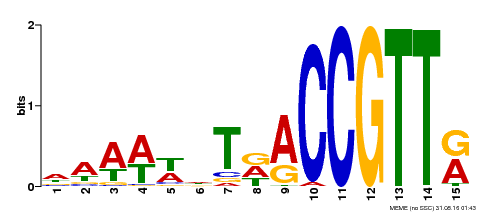
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00482 | DAP | Transfer from AT5G02320 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | GSBRNA2T00069535001 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Slightly induced by ethylene and salicylic acid (SA). {ECO:0000269|PubMed:16463103}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009122788.1 | 0.0 | PREDICTED: myb-related protein 3R-1 isoform X1 | ||||
| Swissprot | Q6R032 | 0.0 | MB3R5_ARATH; Transcription factor MYB3R-5 | ||||
| TrEMBL | A0A397XPD9 | 0.0 | A0A397XPD9_BRACM; Uncharacterized protein | ||||
| STRING | Bra009597.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1667 | 28 | 87 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G02320.2 | 0.0 | myb domain protein 3r-5 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



