 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSBRNA2T00078194001 | ||||||||
| Common Name | GSBRNA2T00078194001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 312aa MW: 35095.5 Da PI: 4.2515 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 57.3 | 2.6e-18 | 62 | 115 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k+++++ +q+++Le+ Fe ++++ e++ +LA++lgL+ rqV vWFqNrRa++k
GSBRNA2T00078194001 62 KKRRLSINQVKALEKNFELENKLEPERKVKLAQELGLQPRQVAVWFQNRRARWK 115
5568999**********************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 129.1 | 1.8e-41 | 61 | 152 | 1 | 92 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLree 90
ekkrrls +qvk+LE++Fe e+kLeperKv+la+eLglqprqvavWFqnrRAR+ktkqlEkdy +Lk++yd+l+++ ++L++++e+L +e
GSBRNA2T00078194001 61 EKKRRLSINQVKALEKNFELENKLEPERKVKLAQELGLQPRQVAVWFQNRRARWKTKQLEKDYGVLKTQYDSLRHNFDSLRRDNESLLQE 150
69*************************************************************************************998 PP
HD-ZIP_I/II 91 lk 92
+
GSBRNA2T00078194001 151 IG 152
85 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50071 | 17.006 | 57 | 117 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 5.6E-17 | 60 | 121 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 5.13E-18 | 60 | 119 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 1.76E-15 | 62 | 118 | No hit | No description |
| Pfam | PF00046 | 1.5E-15 | 62 | 115 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 3.2E-19 | 64 | 124 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 8.7E-6 | 88 | 97 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 92 | 115 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 8.7E-6 | 97 | 113 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 2.0E-17 | 117 | 158 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009788 | Biological Process | negative regulation of abscisic acid-activated signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 312 aa Download sequence Send to blast |
MMKRLSSSDS VGGLISLCPT TSTDQPSPRR YGREFQSMLE GYEEEEEEAV TEERGQTGLA 60 EKKRRLSINQ VKALEKNFEL ENKLEPERKV KLAQELGLQP RQVAVWFQNR RARWKTKQLE 120 KDYGVLKTQY DSLRHNFDSL RRDNESLLQE IGKLKAKLNG EEEVEEDDED EENNAVTMEC 180 DVSVKEEEVS LPEELTDPPS SPPQLLEHSD SFNYRSFTDL RDLLPLKAAA SSVAAAGSSD 240 SSDSSAVLNE ESSSNVTAGP VTVPSGGFLQ FVKMEQTEDH DDFLSGEEAC GFFSDEQPPS 300 LHWYSTVDQW N* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 109 | 117 | RRARWKTKQ |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Bna.2318 | 0.0 | flower| leaf| seed| stem | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Widely expressed. {ECO:0000269|PubMed:10527431, ECO:0000269|PubMed:16055682}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that may act as growth regulators in response to water deficit. Interacts with the core sequence 5'-CAATTATTA-3' of promoters in response to ABA and in an ABI1-dependent manner. Involved in the negative regulation of the ABA signaling pathway. {ECO:0000269|PubMed:10527431, ECO:0000269|PubMed:12065416}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00271 | DAP | Transfer from AT2G22430 | Download |
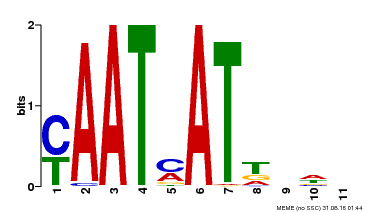
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | GSBRNA2T00078194001 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By water deficit, by abscisic acid (ABA) and by salt stress. Self expression regulation. {ECO:0000269|PubMed:10527431, ECO:0000269|PubMed:12065416, ECO:0000269|PubMed:16055682}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AF268422 | 0.0 | AF268422.1 Brassica rapa subsp. pekinensis hb-6-like protein mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009112528.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein ATHB-6 | ||||
| Swissprot | P46668 | 1e-166 | ATHB6_ARATH; Homeobox-leucine zipper protein ATHB-6 | ||||
| TrEMBL | A0A078FKS5 | 0.0 | A0A078FKS5_BRANA; BnaA09g42630D protein | ||||
| TrEMBL | M4FBQ7 | 0.0 | M4FBQ7_BRARP; Uncharacterized protein | ||||
| STRING | Bra038523.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM545 | 28 | 143 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G22430.1 | 1e-147 | homeobox protein 6 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



