 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSBRNA2T00078905001 | ||||||||
| Common Name | GSBRNA2T00078905001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 548aa MW: 60007.9 Da PI: 7.401 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 97.9 | 6.5e-31 | 209 | 266 | 2 | 60 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS-- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhek 60
dDgynWrKYGqK+vkgsefprsYY+Ct+++C+vkk e+s+ d+++++i+Y+g+H+h+k
GSBRNA2T00078905001 209 DDGYNWRKYGQKHVKGSEFPRSYYKCTHPNCEVKKLFETSH-DGRITDIIYKGTHDHPK 266
8***************************************9.***************85 PP
| |||||||
| 2 | WRKY | 107.2 | 8.2e-34 | 377 | 435 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+v+g+++prsYY+Ct +gCpv+k+ver+++dpk+v++tYeg+Hnh+
GSBRNA2T00078905001 377 LDDGYRWRKYGQKVVRGNPNPRSYYKCTAPGCPVRKHVERASHDPKAVITTYEGKHNHD 435
59********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50811 | 22.905 | 203 | 267 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 2.62E-24 | 206 | 267 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 2.8E-26 | 207 | 267 | IPR003657 | WRKY domain |
| SMART | SM00774 | 7.3E-33 | 208 | 266 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.7E-23 | 209 | 265 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 1.1E-37 | 362 | 437 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.7E-29 | 369 | 437 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 38.629 | 372 | 437 | IPR003657 | WRKY domain |
| SMART | SM00774 | 3.1E-39 | 377 | 436 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 4.4E-26 | 378 | 435 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009961 | Biological Process | response to 1-aminocyclopropane-1-carboxylic acid | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 548 aa Download sequence Send to blast |
MDLPQANNDR EELQVDCSAT VDPTARHNSA GGGGGGGARY KLMSPAMLPI SRSTTNITIP 60 PGLSPTSFLE SPVFISNIKP EPSPTTGSLF KPRPVHVSSS SYTGRAFHQN TFTEHNSSEF 120 EFRPPASNMV YAELDKHKRE PPVQFQAQGH GASHSPEATA SSSEPSRPTP PVQTPPTSSD 180 IPAGSDQEES VQTSQNEPRG SAPPTVLADD GYNWRKYGQK HVKGSEFPRS YYKCTHPNCE 240 VKKLFETSHD GRITDIIYKG THDHPKPQPG RRNSGGVGMA AQEERVGKYP PMTRRDGEKG 300 VYSLSQAIEQ TGNAEVASTT NDGGDVAASN RNKDDPEDDD PYTKRRRLDG NMEITPLVKP 360 IREPRVVVQT LSEVDILDDG YRWRKYGQKV VRGNPNPRSY YKCTAPGCPV RKHVERASHD 420 PKAVITTYEG KHNHDVPTSK SGSSNHHEIQ PRFRPPDETD TISLNLGVGI SSNGPNENQQ 480 LVNQTHPNGV GFRFVHAAPV STYYASMNQY GPRETQSETQ NGDISSLNHS SSYPYPHNNI 540 GRIQTGP* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 1e-37 | 207 | 437 | 8 | 77 | Probable WRKY transcription factor 4 |
| 2lex_A | 1e-37 | 207 | 437 | 8 | 77 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 343 | 347 | KRRRL |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Bna.7158 | 0.0 | seed | ||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element. {ECO:0000250|UniProtKB:Q9SI37}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00455 | DAP | Transfer from AT4G26640 | Download |
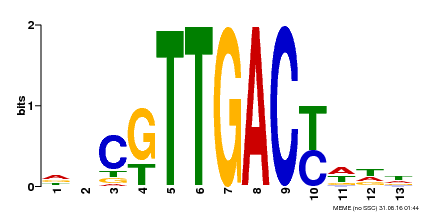
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | GSBRNA2T00078905001 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK352917 | 0.0 | AK352917.1 Thellungiella halophila mRNA, complete cds, clone: RTFL01-11-P10. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009109173.1 | 0.0 | PREDICTED: probable WRKY transcription factor 20 | ||||
| Swissprot | Q93WV0 | 0.0 | WRK20_ARATH; Probable WRKY transcription factor 20 | ||||
| TrEMBL | A0A078FN36 | 0.0 | A0A078FN36_BRANA; BnaA08g14350D protein | ||||
| STRING | Bra010431.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3133 | 28 | 67 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G26640.2 | 0.0 | WRKY family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



