 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSBRNA2T00079331001 | ||||||||
| Common Name | GSBRNA2T00079331001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 455aa MW: 51204.1 Da PI: 6.932 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 45.8 | 1.5e-14 | 190 | 249 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrd.pse.ng..kr.krfslgkfgtaeeAakaaiaarkkleg 55
s y+GV++++++gr++A+++d + g ++ ++++lg ++ +++Aa+a++ a++k++g
GSBRNA2T00079331001 190 SIYRGVTRHRWTGRYEAHLWDnSCRrEGqaRKgRQVYLGGYDKEDKAARAYDLAALKYWG 249
57*******************666664478446**********99*************98 PP
| |||||||
| 2 | AP2 | 49.3 | 1.2e-15 | 294 | 343 | 3 | 55 |
AP2 3 ykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
y+GV++++++grW A+I + +k +lg+f t+eeAa+a++ a+ k++g
GSBRNA2T00079331001 294 YRGVTRHHQQGRWQARIGRVAG---NKDLYLGTFATEEEAAEAYDIAAIKFRG 343
9***************988532...5************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 1.8E-11 | 190 | 249 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 2.34E-21 | 190 | 259 | No hit | No description |
| SuperFamily | SSF54171 | 2.09E-16 | 190 | 258 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 2.5E-26 | 191 | 263 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 18.755 | 191 | 257 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 7.0E-15 | 191 | 257 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.4E-6 | 192 | 203 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 9.65E-11 | 292 | 351 | No hit | No description |
| SuperFamily | SSF54171 | 9.15E-18 | 292 | 352 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 3.3E-30 | 293 | 357 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 19.269 | 293 | 351 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 3.5E-18 | 293 | 351 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 5.9E-10 | 294 | 343 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.4E-6 | 333 | 353 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010492 | Biological Process | maintenance of shoot apical meristem identity | ||||
| GO:0060772 | Biological Process | leaf phyllotactic patterning | ||||
| GO:0060774 | Biological Process | auxin mediated signaling pathway involved in phyllotactic patterning | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 455 aa Download sequence Send to blast |
MANWLTFALS PMEMMKSPDH THFVSYDDSS TPYLIDNLYV LKEEAETSMA DSTNLASFFN 60 PQTHSPTHIP KLEDFLGDSS SFVRFPDNQP DTLDSSSLTQ IYDPRHLTGV TGLFSDRQHD 120 FKAVEGGVSE GCTKEGALSL AVNNTDGERV RNSRKVKVSK KQAKAVETTS TDDLTKKNKK 180 VAESFGQRTS IYRGVTRHRW TGRYEAHLWD NSCRREGQAR KGRQVYLGGY DKEDKAARAY 240 DLAALKYWGP AATTNFQIAT YSKELEEMNH MTKQEFIASI RRKSSGFSRG ASMYRGVTRH 300 HQQGRWQARI GRVAGNKDLY LGTFATEEEA AEAYDIAAIK FRGINAVTNF EMNRYDVEAI 360 MNSSFPVGGS AVKRHKQLSL ESPPPLPPSD DDHNIQQLLL PSSSVELDPN SIPCGIPFDP 420 SVLYLHQNFF QHYPDPTVPM NQADQFFMWS NQSY* |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Confined to the central region of inflorescence ane floral meristems, progressively restricted to the innermost cells of the dome. Also detected in developing stamen locules and later in sporogonous cells within locules. In carpel primordia, found in placenta and in young ovule primordia. {ECO:0000269|PubMed:15988559}. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in roots, seedlings, inflorescence, and siliques. Also detected at low levels in leaves. {ECO:0000269|PubMed:15988559}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00589 | DAP | Transfer from AT5G65510 | Download |
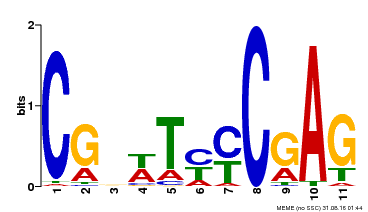
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | GSBRNA2T00079331001 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK353404 | 0.0 | AK353404.1 Thellungiella halophila mRNA, complete cds, clone: RTFL01-30-G24. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013628400.1 | 0.0 | PREDICTED: AP2-like ethylene-responsive transcription factor AIL7 | ||||
| Swissprot | Q6J9N8 | 0.0 | AIL7_ARATH; AP2-like ethylene-responsive transcription factor AIL7 | ||||
| TrEMBL | A0A078FNZ3 | 0.0 | A0A078FNZ3_BRANA; BnaC03g48820D protein | ||||
| STRING | Bo3g099090.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3333 | 27 | 65 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G65510.1 | 0.0 | AINTEGUMENTA-like 7 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



