 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSBRNA2T00098926001 | ||||||||
| Common Name | GSBRNA2T00098926001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 312aa MW: 36227.9 Da PI: 5.9058 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 178.5 | 1.7e-55 | 6 | 133 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLp.k.kvkaeekewyfFskrdkkyatgkrknratksgyWkatgk 88
lppGfrFhPtdeelv +yL +kv+g+++el e+i+evd+yk+ePwdLp k + +++ ewyf+s+rdkky++g+r+nrat++gyWkatgk
GSBRNA2T00098926001 6 LPPGFRFHPTDEELVAYYLDRKVNGRTIEL-EIIPEVDLYKCEPWDLPeKsFLPGNDMEWYFYSTRDKKYPNGSRTNRATRAGYWKATGK 94
79****************************.99**************95334455788******************************** PP
NAM 89 dkevlskkgelvglkktLvfykgrapkgektdWvmheyrl 128
d++v s k+ + g+kktLv+y+grap+g +t+Wvmheyrl
GSBRNA2T00098926001 95 DRAVES-KKMKLGMKKTLVYYRGRAPHGLRTNWVMHEYRL 133
*****9.8999***************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 1.44E-62 | 3 | 157 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 59.279 | 6 | 157 | IPR003441 | NAC domain |
| Pfam | PF02365 | 6.9E-29 | 7 | 133 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 312 aa Download sequence Send to blast |
MAPMSLPPGF RFHPTDEELV AYYLDRKVNG RTIELEIIPE VDLYKCEPWD LPEKSFLPGN 60 DMEWYFYSTR DKKYPNGSRT NRATRAGYWK ATGKDRAVES KKMKLGMKKT LVYYRGRAPH 120 GLRTNWVMHE YRLTHLPFSS PSSSIKESYA LCRVFKKHIQ IPKRKDEEMM MGTSVGKEKK 180 EEEEENMWRK CDKVMMERES DEDESLKIAS AETSSSELTQ GILLDEANNS SNFPLHFSSS 240 LLDDHDQFFA NYSHLPYYPP LQLQDFPQIS INEAQIMSTE YSKQQDFQCR DSMNGTLDEI 300 FSFSSSATFP L* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 2e-55 | 1 | 158 | 12 | 166 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 2e-55 | 1 | 158 | 12 | 166 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 2e-55 | 1 | 158 | 12 | 166 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 2e-55 | 1 | 158 | 12 | 166 | NO APICAL MERISTEM PROTEIN |
| 3swm_A | 1e-55 | 1 | 158 | 15 | 169 | NAC domain-containing protein 19 |
| 3swm_B | 1e-55 | 1 | 158 | 15 | 169 | NAC domain-containing protein 19 |
| 3swm_C | 1e-55 | 1 | 158 | 15 | 169 | NAC domain-containing protein 19 |
| 3swm_D | 1e-55 | 1 | 158 | 15 | 169 | NAC domain-containing protein 19 |
| 3swp_A | 1e-55 | 1 | 158 | 15 | 169 | NAC domain-containing protein 19 |
| 3swp_B | 1e-55 | 1 | 158 | 15 | 169 | NAC domain-containing protein 19 |
| 3swp_C | 1e-55 | 1 | 158 | 15 | 169 | NAC domain-containing protein 19 |
| 3swp_D | 1e-55 | 1 | 158 | 15 | 169 | NAC domain-containing protein 19 |
| 4dul_A | 2e-55 | 1 | 158 | 12 | 166 | NAC domain-containing protein 19 |
| 4dul_B | 2e-55 | 1 | 158 | 12 | 166 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in a few sieve element cells before enucleation and in phloem-pole pericycle cells. {ECO:0000269|PubMed:25081480}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor directing sieve element enucleation and cytosol degradation. Not required for formation of lytic vacuoles. Regulates, with NAC045, the transcription of NEN1, NEN2, NEN3, NEN4, RTM1, RTM2, UBP16, PLDZETA, ABCB10 and At1g26450. {ECO:0000269|PubMed:25081480}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00205 | DAP | Transfer from AT1G54330 | Download |
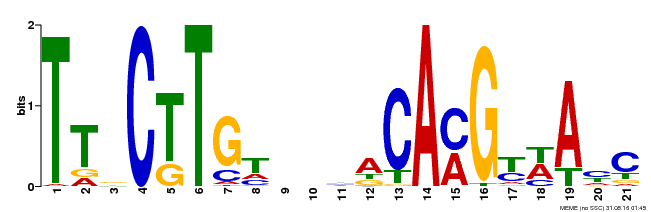
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | GSBRNA2T00098926001 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK176278 | 1e-147 | AK176278.1 Arabidopsis thaliana mRNA for hypothetical protein, complete cds, clone: RAFL23-20-J17. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_022572074.1 | 0.0 | NAC domain-containing protein 71-like | ||||
| Swissprot | Q9FFI5 | 4e-82 | NAC86_ARATH; NAC domain-containing protein 86 | ||||
| TrEMBL | A0A3P6BE60 | 0.0 | A0A3P6BE60_BRACM; Uncharacterized protein | ||||
| STRING | Bo6g036040.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM8123 | 26 | 40 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G54330.1 | 1e-170 | NAC domain containing protein 20 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



