 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSBRNA2T00101975001 | ||||||||
| Common Name | GSBRNA2T00101975001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | YABBY | ||||||||
| Protein Properties | Length: 181aa MW: 19745.5 Da PI: 10.157 | ||||||||
| Description | YABBY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | YABBY | 142.2 | 5.8e-44 | 14 | 156 | 4 | 163 |
YABBY 4 fssseqvCyvqCnfCntilavsvPstslfkvvtvrCGhCtsllsvnlakasqllaaeshldeslkeelleelkveeenlksnvekeesas 93
++e++ yv+C+ Cntilav +P++ ++ +vtv+CGhC +l ++ + + + h + +l++ + ++ + k+ s+s
GSBRNA2T00101975001 14 SPQAEHLYYVRCSICNTILAVGIPMKRMLDTVTVKCGHCGNLSFLTTT-----PPLQGH----VSLTLQQMQ-----SFGGSEYKKGSSS 89
5689************************************97553332.....233333....223333333.....3444444445544 PP
YABBY 94 tsvsseklsenedeevprvppvirPPekrqrvPsaynrfikeeiqrikasnPdishreafsaaaknWahf 163
+s ss +++++ +pr+p v++PPek+qr Psaynrf+++eiqrik++nP+i hreafsaaaknWa +
GSBRNA2T00101975001 90 SSSSS---TSSDQPPSPRPPFVVKPPEKKQRLPSAYNRFMRDEIQRIKSANPEIPHREAFSAAAKNWAKY 156
44444...3467788999999***********************************************86 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04690 | 2.3E-50 | 17 | 156 | IPR006780 | YABBY protein |
| SuperFamily | SSF47095 | 1.26E-8 | 100 | 155 | IPR009071 | High mobility group box domain |
| Gene3D | G3DSA:1.10.30.10 | 7.0E-5 | 110 | 155 | IPR009071 | High mobility group box domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010254 | Biological Process | nectary development | ||||
| GO:0010582 | Biological Process | floral meristem determinacy | ||||
| GO:0048479 | Biological Process | style development | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 181 aa Download sequence Send to blast |
MNLEEKPTMA SRVSPQAEHL YYVRCSICNT ILAVGIPMKR MLDTVTVKCG HCGNLSFLTT 60 TPPLQGHVSL TLQQMQSFGG SEYKKGSSSS SSSSTSSDQP PSPRPPFVVK PPEKKQRLPS 120 AYNRFMRDEI QRIKSANPEI PHREAFSAAA KNWAKYIPNS PTSITSGASN IHGFGFGEKK 180 * |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: In carpels, expression starts when the gynoecial primordia becomes distinct. First expressed in the lateral region of each carpel. Later resolves into two distinct domains, epidermal and internal. In epidermal tissues, mostly localized on the outer surface, successively from the base to the tip of the gynoecium and finally confined to the valve region before disappearing during last flowering stages. In internal tissues, first confined to four discrete zones adjacent to the future placental tissue occupying the full length of the elongating cylinder, declining from the apical regions and disappearing after the ovule primordia arise. Before nectaries initiation, expression occupies a ring of receptacle cells between the stamen and sepal primordia, from where nectaries will develop. Strongly expressed in nectaries until latest flowering stages. {ECO:0000269|PubMed:10225998}. | |||||
| Uniprot | TISSUE SPECIFICITY: Restricted to flowers, mostly in carpels and nectaries. Expressed at low levels in sepal primordia (buds), sepal receptacle and developing petal. Not detected in placental tissues, septum, stigma and ovules. {ECO:0000269|PubMed:10225998}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor required for the initiation of nectary development. Also involved in suppressing early radial growth of the gynoecium, in promoting its later elongation and in fusion of its carpels by regulating both cell division and expansion. Establishes the polar differentiation in the carpels by specifying abaxial cell fate in the ovary wall. Regulates both cell division and expansion. {ECO:0000269|PubMed:10225997, ECO:0000269|PubMed:10225998, ECO:0000269|PubMed:10535738, ECO:0000269|PubMed:11714690, ECO:0000269|PubMed:15598802, ECO:0000269|Ref.10}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00220 | DAP | Transfer from AT1G69180 | Download |
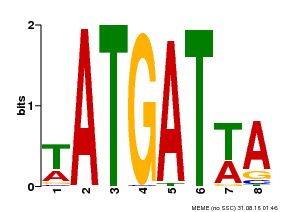
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | GSBRNA2T00101975001 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated by SPT and by A class genes AP2 and LUG in the outer whorl. In the third whorl, B class genes AP3 and PI, and the C class gene AG act redundantly with each other and in combination with SEP1, SEP2, SEP3, SHP1 and SHP2 to activate CRC in nectaries and carpels. LFY enhances its expression. {ECO:0000269|PubMed:10225998, ECO:0000269|PubMed:15598802}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | DQ119055 | 0.0 | DQ119055.1 Brassica juncea transcription factor CRC mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009105464.1 | 1e-122 | PREDICTED: protein CRABS CLAW | ||||
| Refseq | XP_013652319.1 | 1e-122 | protein CRABS CLAW | ||||
| Swissprot | Q8L925 | 1e-107 | CRC_ARATH; Protein CRABS CLAW | ||||
| TrEMBL | A0A3P6BLF5 | 1e-131 | A0A3P6BLF5_BRACM; Uncharacterized protein | ||||
| STRING | Bo6g108630.1 | 1e-118 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM9725 | 28 | 35 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G69180.1 | 1e-108 | YABBY family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



