 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSBRNA2T00103449001 | ||||||||
| Common Name | GSBRNA2T00103449001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | EIL | ||||||||
| Protein Properties | Length: 586aa MW: 65727.1 Da PI: 5.0028 | ||||||||
| Description | EIL family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | EIN3 | 433.9 | 1.5e-132 | 41 | 365 | 1 | 324 |
XXXXXXXXXXXXXXXXXXXXXXX..XXXXX.XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX CS
EIN3 1 eelkkrmwkdqmllkrlkerkkqlledkeaatgakksnksneqarrkkmsraQDgiLkYMlkemevcnaqGfvYgiipekgkpvegasds 90
e+l++rmwkd+++lkr+ker+k + + ++ +k+++qa+rkkmsraQDgiLkYMlk mevc+++GfvYgiipekgkpv+g+sd+
GSBRNA2T00103449001 41 EDLERRMWKDRVRLKRIKERQKGDS--QGP-QAKEPPKKISDQAQRKKMSRAQDGILKYMLKLMEVCKVRGFVYGIIPEKGKPVSGSSDN 127
89********************965..555.4456689**************************************************** PP
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX....XX----STTS-HHHHHHHHHHHSSSSSS-TTS--TTT--HHHH---S--HH CS
EIN3 91 LraWWkekvefdrngpaaiskyqaknlilsgesslqtersseshslselqDTtlgSLLsalmqhcdppqrrfplekgvepPWWPtGkelw 180
+raWWkekv+fd+ngpaai+ky+ ++l++++++++ ++++ l++lqD+tlgSLLs+lmqhcdppqr++plekg++pPWWPtGke+w
GSBRNA2T00103449001 128 IRAWWKEKVKFDKNGPAAIAKYEEECLAFGKSDGN----RNSQFVLQDLQDATLGSLLSSLMQHCDPPQRKYPLEKGTPPPWWPTGKEEW 213
*****************************987777....77999********************************************** PP
HHHHT--TT--.-----GGG--HHHHHHHHHHHHHHTGGGHHHHHHTTTTSSSSTTT--SHHHHHHHHHHTTTTT-S--XXXX...XX.. CS
EIN3 181 wgelglskdqgtppykkphdlkkawkvsvLtavikhmsptieeirelerqskylqdkmsakesfallsvlnqeekecatvsah...ss.. 265
w +lgl+++q+ ppy+kphdlkk+wkv+vLtavi+hmsp+i++i++++rqsk+lqdkm+akes+++l+vlnqee+++++ s++ s+
GSBRNA2T00103449001 214 WVKLGLPQSQS-PPYRKPHDLKKMWKVGVLTAVINHMSPDIAKIKRHVRQSKCLQDKMTAKESAIWLAVLNQEESLIQQPSSDngtSNvt 302
**********9.9**********************************************************************6542278 PP
...XXXX....XXXXXXXXXXXXXXXXXXXXXX.XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX. CS
EIN3 266 ...slrk....qspkvtlsceqkedvegkkeskikhvqavkttagfpvvrkrkkkpsesakvsske 324
+ +++k+++++++++dv+g++e + + ++++++++++p++ ++ + +++ k+ +++
GSBRNA2T00103449001 303 ethR--RgnnaDRRKTVINSDSDYDVDGTEEASGSVSSKDSRRNQVPAATSQQ-PVRDQDKAGKHK 365
7540..33445888999**********99999999999999999555555544.333333333332 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04873 | 1.0E-125 | 41 | 286 | No hit | No description |
| Gene3D | G3DSA:1.10.3180.10 | 3.7E-71 | 162 | 294 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| SuperFamily | SSF116768 | 8.5E-61 | 167 | 289 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 586 aa Download sequence Send to blast |
MGGGDLALSV ADIRMENEQP DDLASDTVAE IDVSDEEIDA EDLERRMWKD RVRLKRIKER 60 QKGDSQGPQA KEPPKKISDQ AQRKKMSRAQ DGILKYMLKL MEVCKVRGFV YGIIPEKGKP 120 VSGSSDNIRA WWKEKVKFDK NGPAAIAKYE EECLAFGKSD GNRNSQFVLQ DLQDATLGSL 180 LSSLMQHCDP PQRKYPLEKG TPPPWWPTGK EEWWVKLGLP QSQSPPYRKP HDLKKMWKVG 240 VLTAVINHMS PDIAKIKRHV RQSKCLQDKM TAKESAIWLA VLNQEESLIQ QPSSDNGTSN 300 VTETHRRGNN ADRRKTVINS DSDYDVDGTE EASGSVSSKD SRRNQVPAAT SQQPVRDQDK 360 AGKHKRRKRP RIRSGTLNVQ DEEQVEAEGR NVLPDMNHVE APMLDYNING TTNHHEEGVL 420 EPNISLGPEE NGLELVVPEF DSNYTYLPPV DGQAMMPVDE RPMLYGANPN QELQFGSGYN 480 YYNTSAVFVH NQEEDLIHTQ IEMNSQAPPH SNGFDGQGGV LQPHGNEEVG VAGRDMPPQF 540 QSDQDKLLDS NILSPFNDLP FDSSTFYSGF DSFGAFDDDY SWFGA* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wij_A | 1e-84 | 166 | 297 | 9 | 140 | ETHYLENE-INSENSITIVE3-like 3 protein |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may be involved in the ethylene response pathway. {ECO:0000269|PubMed:9215635, ECO:0000269|PubMed:9851977}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00649 | PBM | Transfer from LOC_Os09g31400 | Download |
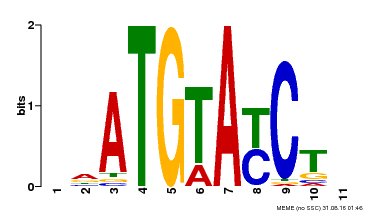
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | GSBRNA2T00103449001 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013592375.1 | 0.0 | PREDICTED: ETHYLENE INSENSITIVE 3-like 3 protein isoform X1 | ||||
| Swissprot | O23116 | 0.0 | EIL3_ARATH; ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| TrEMBL | A0A0D3D0F8 | 0.0 | A0A0D3D0F8_BRAOL; Uncharacterized protein | ||||
| STRING | Bo6g118030.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4658 | 25 | 48 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73730.1 | 0.0 | ETHYLENE-INSENSITIVE3-like 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



