 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSBRNA2T00105785001 | ||||||||
| Common Name | GSBRNA2T00105785001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | YABBY | ||||||||
| Protein Properties | Length: 245aa MW: 27620.8 Da PI: 8.377 | ||||||||
| Description | YABBY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | YABBY | 236.8 | 4.9e-73 | 17 | 205 | 2 | 170 |
YABBY 2 dvfssseqvCyvqCnfCntilavsvPstslfkvvtvrCGhCtsllsvnlakasqllaaeshldeslkee.......lleelkveeenlks 84
++ s se++CyvqCnfC+tilavsvP+tslfk+vtvrCG+C++lls ++++s +l+a+++++ +l ++ +leel+ + +n+++
GSBRNA2T00105785001 17 EHLSPSEHLCYVQCNFCETILAVSVPYTSLFKTVTVRCGCCANLLS--VNMRSLVLPASNQIQLQLGPQsyftpqnILEELREAPSNMNM 104
67899*****************************************..5678899***********999999999*************** PP
YABBY 85 nvekeesastsvsseklsenedeevprvppvir.......P.........PekrqrvPsaynrfikeeiqrikasnPdishreafsaaak 158
++++++ ++++++s++ + ++++e+p++pp++r P PekrqrvPsaynrfikeeiqrika+nPdishreafsaaak
GSBRNA2T00105785001 105 MMINQHPNMNDIPSFM-DLHQKHEIPKAPPINRhikrwpyPkkndevmmaPEKRQRVPSAYNRFIKEEIQRIKAGNPDISHREAFSAAAK 193
**************65.9999999999999887555554414555555559*************************************** PP
YABBY 159 nWahfPkihfgl 170
nWahfP+ihfgl
GSBRNA2T00105785001 194 NWAHFPHIHFGL 205
**********97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04690 | 6.3E-66 | 21 | 205 | IPR006780 | YABBY protein |
| SuperFamily | SSF47095 | 3.8E-8 | 141 | 198 | IPR009071 | High mobility group box domain |
| Gene3D | G3DSA:1.10.30.10 | 8.8E-5 | 153 | 199 | IPR009071 | High mobility group box domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009909 | Biological Process | regulation of flower development | ||||
| GO:0009933 | Biological Process | meristem structural organization | ||||
| GO:0009944 | Biological Process | polarity specification of adaxial/abaxial axis | ||||
| GO:0010093 | Biological Process | specification of floral organ identity | ||||
| GO:0010154 | Biological Process | fruit development | ||||
| GO:0010158 | Biological Process | abaxial cell fate specification | ||||
| GO:0010159 | Biological Process | specification of organ position | ||||
| GO:0010450 | Biological Process | inflorescence meristem growth | ||||
| GO:1902183 | Biological Process | regulation of shoot apical meristem development | ||||
| GO:2000024 | Biological Process | regulation of leaf development | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 245 aa Download sequence Send to blast |
MSMSSMSSPS SAVFSAEHLS PSEHLCYVQC NFCETILAVS VPYTSLFKTV TVRCGCCANL 60 LSVNMRSLVL PASNQIQLQL GPQSYFTPQN ILEELREAPS NMNMMMINQH PNMNDIPSFM 120 DLHQKHEIPK APPINRHIKR WPYPKKNDEV MMAPEKRQRV PSAYNRFIKE EIQRIKAGNP 180 DISHREAFSA AAKNWAHFPH IHFGLMPDNQ PVKKTNMPQQ AGEENMGMKE GFYAPAANVG 240 MTPY* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Bna.17602 | 1e-132 | flower| seed | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Expressed in subepidermal cells of anlagen regions, then in abaxial part of primordia and finally in differentiating organs. Levels decrease in differentiated organs. In embryo the expression starts during the transition between globular and heart stages in the cotyledon anlagen. From the heart stage, expression expands to abaxial domain of the cotyledon primordia and decrease as the embryo matures. In stamen, expression restricted to the abaxial region differentiating into the connective. In gynoecium, expressed in the abaxial cell layers differentiating into the valves. {ECO:0000269|PubMed:10457020}. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in abaxial regions of young lateral aerial organs, and in primordia leading to cotyledons, leaves, flower meristems, sepals, petals, stamen and carpels, but not in roots. {ECO:0000269|PubMed:10323860, ECO:0000269|PubMed:10457020, ECO:0000269|PubMed:14555697, ECO:0000269|Ref.3}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Involved in the abaxial cell fate determination during embryogenesis and organogenesis. Regulates the initiation of embryonic shoot apical meristem (SAM) development (PubMed:10323860, PubMed:10331982, PubMed:10457020, PubMed:11812777, PubMed:12417699, PubMed:9878633, Ref.3, Ref.6, PubMed:19837869). Required during flower formation and development, particularly for the patterning of floral organs. Positive regulator of class B (AP3 and PI) activity in whorls 2 and 3. Negative regulator of class B activity in whorl 1 and of SUP activity in whorl 3. Interacts with class A proteins (AP1, AP2 and LUG) to repress class C (AG) activity in whorls 1 and 2. Contributes to the repression of KNOX genes (STM, KNAT1/BP and KNAT2) to avoid ectopic meristems. Binds DNA without sequence specificity. In vitro, can compete and displace the AP1 protein binding to DNA containing CArG box (PubMed:10323860, PubMed:10331982, PubMed:10457020, PubMed:11812777, PubMed:12417699, PubMed:9878633, Ref.3, Ref.6). {ECO:0000269|PubMed:10323860, ECO:0000269|PubMed:10331982, ECO:0000269|PubMed:10457020, ECO:0000269|PubMed:11812777, ECO:0000269|PubMed:12417699, ECO:0000269|PubMed:19837869, ECO:0000269|PubMed:9878633, ECO:0000269|Ref.3, ECO:0000269|Ref.6}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00620 | PBM | Transfer from AT2G45190 | Download |
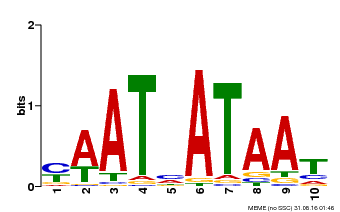
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | GSBRNA2T00105785001 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | JQ234919 | 1e-151 | JQ234919.1 Brassica rapa subsp. pekinensis clone 5-702 putative male sterile protein mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013728413.1 | 1e-158 | axial regulator YABBY 1 isoform X1 | ||||
| Swissprot | O22152 | 1e-135 | YAB1_ARATH; Axial regulator YABBY 1 | ||||
| TrEMBL | A0A3P6G8H8 | 0.0 | A0A3P6G8H8_BRAOL; Uncharacterized protein | ||||
| STRING | Bo6g073320.1 | 1e-156 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2217 | 28 | 77 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G45190.1 | 1e-136 | YABBY family protein | ||||



