 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSBRNA2T00113560001 | ||||||||
| Common Name | GSBRNA2T00113560001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 729aa MW: 83772.6 Da PI: 9.8507 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 48.4 | 1.7e-15 | 538 | 583 | 5 | 55 |
HHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 5 hnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hne ErrRRdriN+++ +L++llP+a K +K++iL ++e++k+Lq
GSBRNA2T00113560001 538 HNESERRRRDRINQRMRTLQKLLPTA-----SKADKVSILDDVIEHLKQLQ 583
*************************9.....7******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03763 | 2.8E-19 | 192 | 291 | IPR005516 | Remorin, C-terminal |
| PROSITE profile | PS50888 | 17.06 | 533 | 582 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 7.33E-18 | 536 | 604 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 7.4E-17 | 536 | 598 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 2.89E-7 | 538 | 587 | No hit | No description |
| Pfam | PF00010 | 9.4E-13 | 538 | 583 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.9E-16 | 539 | 588 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0016021 | Cellular Component | integral component of membrane | ||||
| GO:0016607 | Cellular Component | nuclear speck | ||||
| GO:0003690 | Molecular Function | double-stranded DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 729 aa Download sequence Send to blast |
MFLYSGIRIL AIFYNIGRIY EAQTILESLG ENLVKQRRRR VSVSENKQTD MPNLSELAIT 60 DNPALNWLKN QAYWYEKMDY YDEKESEFAT FFFFFFFFCF FFFFFFFFVF SMEEAYKENA 120 KRMREETKRS RKKKTNPVIA KSEVKRINRS FTQDLTIGEE SFKKQQMDNP QKDRREQEIG 180 SSSRTLGLAS APSKADSWEK SQLDKIKLRY EKMKAEIVGW ENERKSAAKL RMEKRKSELQ 240 KRTEINNQHY KTKLARIQVI ADGAKKQLEE KKRSQEAQVQ EKVKKMRRTG KREDSRQGNS 300 NPLPHMCFLH IKTIVIAGTI VSLLYYFNVD QMKIHYMSCR CDCEELENRG GHQDYTTKTK 360 KKKRKAELSN YGVKELTWEN GQLTVHGLGE GVEPTTTSAN LLWTQALNGC ETLESVVHQA 420 ALQPSKLQSQ NGRDHNNSES KDGSCSRKRG YPQEMDCWFS GQEESHRVGH SVTASASGTN 480 MSWASFESGR SLKTARTGDR DYLFSGSETQ ETEGDEQETR GEAGRSNNGR RGRAAAIHNE 540 SERRRRDRIN QRMRTLQKLL PTASKADKVS ILDDVIEHLK QLQAQVQFMS LRANLPQQMM 600 MPQLPPPQSV LSIQHQQQQQ QQQQQQQQQF QMSLLATMAR MGMGGGGNAY GGLVPPHPPL 660 MVPPMDNRDC TNASSASLTD PYSAFLAQTM NMDLYNKMAA AIYRQQSDQT PKVNTGMPSS 720 SSNHEKRD* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 359 | 364 | KKKKRK |
| 2 | 541 | 546 | ERRRRD |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Barely detectable in dark-grown seedlings. Induced after 2 hours of light exposure. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in flowers. {ECO:0000269|PubMed:12679534}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor acting negatively in the phytochrome B signaling pathway under prolonged red light. Regulates PHYB abundance at the post-transcriptional level, possibly via the ubiquitin-proteasome pathway. May regulate the expression of a subset of genes by binding to the G-box motif. {ECO:0000269|PubMed:15486100, ECO:0000269|PubMed:18252845}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00571 | DAP | Transfer from AT5G61270 | Download |
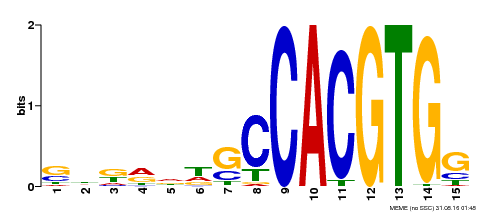
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | GSBRNA2T00113560001 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By red light. Stable upon light exposure. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK352734 | 0.0 | AK352734.1 Thellungiella halophila mRNA, complete cds, clone: RTFL01-03-L18. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013736938.1 | 0.0 | transcription factor PIF7 | ||||
| Swissprot | Q570R7 | 0.0 | PIF7_ARATH; Transcription factor PIF7 | ||||
| TrEMBL | A0A3P5ZR11 | 0.0 | A0A3P5ZR11_BRACM; Uncharacterized protein | ||||
| STRING | Bra012972.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM14123 | 18 | 22 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G61270.1 | 1e-102 | phytochrome-interacting factor7 | ||||



