 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSBRNA2T00117117001 | ||||||||
| Common Name | GSBRNA2T00117117001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | E2F/DP | ||||||||
| Protein Properties | Length: 535aa MW: 58032 Da PI: 4.7619 | ||||||||
| Description | E2F/DP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | E2F_TDP | 82 | 5e-26 | 213 | 278 | 1 | 71 |
E2F_TDP 1 rkeksLrlltqkflkllekseegivtlnevakeLvsedvknkrRRiYDilNVLealnliekkekneirwkg 71
r+++sL+llt+kf++l++++++g+++ln++a++L +v ++RRiYDi+NVLe+++liek kn+i wkg
GSBRNA2T00117117001 213 RYDSSLGLLTKKFVNLIKQAKDGMLDLNKAAETL---EV--QKRRIYDITNVLEGIDLIEKPLKNRILWKG 278
6899******************************...99..****************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.10 | 7.4E-28 | 209 | 280 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SMART | SM01372 | 2.9E-32 | 213 | 278 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| SuperFamily | SSF46785 | 2.18E-16 | 213 | 276 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Pfam | PF02319 | 8.8E-23 | 215 | 278 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| SuperFamily | SSF144074 | 9.94E-33 | 291 | 395 | No hit | No description |
| CDD | cd14660 | 2.61E-49 | 291 | 395 | No hit | No description |
| Pfam | PF16421 | 7.1E-33 | 295 | 393 | IPR032198 | E2F transcription factor, CC-MB domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0051446 | Biological Process | positive regulation of meiotic cell cycle | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005667 | Cellular Component | transcription factor complex | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 535 aa Download sequence Send to blast |
MSGGVRSSPG PSRQPPPPPS SSAASVPVVP PISRHLAFAS TKPPFHPSDN YHRFTPSHLT 60 NNLVNGCGGL VDREEDAVVL RSPVSSPRST IYSNCYGLLS SNCAVLISPT VKLCVSQSVS 120 IKVMLHEAWS ILFIYHSRKR KSTMDVVTTT TSSTSNGFTS SDGFTSSPCR TPVSAKGGRV 180 NIKSKSKGNQ SIPQTPISNI GSPVTLTPSG SCRYDSSLGL LTKKFVNLIK QAKDGMLDLN 240 KAAETLEVQK RRIYDITNVL EGIDLIEKPL KNRILWKGVD ASGPGDVDAD VSDVQAEIEN 300 LSLEEQALDN QIRETEQRLR DLSENEKNQK WLFVTEEDIK SLPGFQNQTL IAVKAPHGTT 360 LEVPDPDEGV GHPQRRYRII LRSTMGPIDV YLVSEFEEKF EDTNGPAEPP PPLCLPIASC 420 SGSTENHDIE ALTADNTGTA IEDQLSQVHA HAQPGETSDL NYLEEQVGGM LKITPSDVEN 480 DDTDYWLLSN ADISMTDIWK TDSGIDWDYG IADVSTPPPM IGDISPTAID SIPR* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1cf7_A | 6e-23 | 208 | 279 | 2 | 74 | PROTEIN (TRANSCRIPTION FACTOR E2F-4) |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 136 | 141 | SRKRKS |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Expressed in a cell cycle-dependent manner. Most abundant in early S phase. Decreased expression during the passage into G2. {ECO:0000269|PubMed:11108847, ECO:0000269|PubMed:11669580, ECO:0000269|PubMed:11786543}. | |||||
| Uniprot | TISSUE SPECIFICITY: Highly expressed in the shoot apical meristem, emerging leaf primordia, and vascular tissues of young leaf primordia. Expressed in flowers, in epidermis and cortex of hypocotyls, and at lower levels in leaves. {ECO:0000269|PubMed:11108847, ECO:0000269|PubMed:11889041}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds DNA cooperatively with DP proteins through the E2 recognition site, 5'-TTTC[CG]CGC-3' found in the promoter region of a number of genes whose products are involved in cell cycle regulation or in DNA replication. The binding of retinoblastoma-related proteins represses transactivation. Regulates gene expression both positively and negatively. Activates the expression of E2FB. Involved in the control of cell-cycle progression from G1 to S phase. Stimulates cell proliferation and delays differentiation. {ECO:0000269|PubMed:11669580, ECO:0000269|PubMed:11786543, ECO:0000269|PubMed:11862494, ECO:0000269|PubMed:11889041, ECO:0000269|PubMed:11891240, ECO:0000269|PubMed:12913157, ECO:0000269|PubMed:15377755, ECO:0000269|PubMed:16514015, ECO:0000269|PubMed:19662336}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00293 | DAP | Transfer from AT2G36010 | Download |
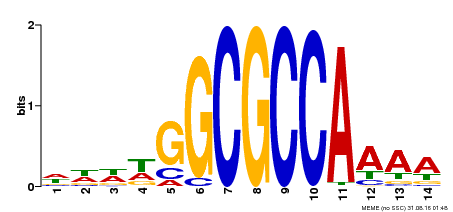
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | GSBRNA2T00117117001 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | JN942984 | 0.0 | JN942984.1 Brassica juncea E2F transcription factor (E2Fa) mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009143621.1 | 0.0 | PREDICTED: transcription factor E2FA-like isoform X1 | ||||
| Swissprot | Q9FNY0 | 0.0 | E2FA_ARATH; Transcription factor E2FA | ||||
| TrEMBL | A0A3P5YSW9 | 0.0 | A0A3P5YSW9_BRACM; Uncharacterized protein | ||||
| STRING | Bra005305.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM808 | 28 | 121 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G36010.1 | 0.0 | E2F transcription factor 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



