 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSBRNA2T00119141001 | ||||||||
| Common Name | GSBRNA2T00119141001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 1033aa MW: 114140 Da PI: 8.3982 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 124.4 | 4.6e-39 | 130 | 207 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqv++C +dls+ k+yhrrhkvCevhska+++lv +++qrfCqqCsrfh lsefDe+krsCrrrLa+hn+rrrk+++
GSBRNA2T00119141001 130 VCQVDNCCQDLSHGKDYHRRHKVCEVHSKATKALVGKQMQRFCQQCSRFHLLSEFDEGKRSCRRRLAGHNRRRRKTTQ 207
6**************************************************************************875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 4.3E-32 | 124 | 192 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 31.573 | 128 | 205 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 1.96E-35 | 129 | 207 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 4.4E-27 | 131 | 204 | IPR004333 | Transcription factor, SBP-box |
| CDD | cd00204 | 6.90E-10 | 786 | 920 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 3.6E-8 | 786 | 934 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 2.13E-8 | 788 | 924 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 10.445 | 829 | 939 | IPR020683 | Ankyrin repeat-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0016021 | Cellular Component | integral component of membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1033 aa Download sequence Send to blast |
MEEVGAQVAA PLFIHQSLSP MGRKRNLYHH QMPNRLVPPS QPRDEWNSNL WDWDSRRFEA 60 KPVDAEPLRL GSQSQSQSQS QTQFDLTSRK VGEERGLDLN LGSCLNAAEA ARPSKKGRSG 120 SPGTGGNYPV CQVDNCCQDL SHGKDYHRRH KVCEVHSKAT KALVGKQMQR FCQQCSRFHL 180 LSEFDEGKRS CRRRLAGHNR RRRKTTQPEE VASGVVAPGN RDSTSNANMD LMALLTALAC 240 AQGKNEVRPM GSPAVPQREQ LLQILNKINA LPLPMDLVSK LNSIGSLARK NTDHPVANPQ 300 NDVNGDSPST MDLLAVLSET LGSSSPDTLA ILSQGGFGTK ENDKAKLSSY DHVATTNVEK 360 RTVGGERSSS SNQSPSQDSD SHAQDTRSSL SLQLFTSSPE DESRRPAVAS SRKYYSSASS 420 NPVEDRSPSS SPVMQELFPL QRSPETMRSK NHNNTSPVRT AGCLPLDLFG TSNRGAANPN 480 FKGFGQQCGY ASSGSDYSPP SLNSDAQDRT GKIVFKLLDK DPSQLPGTLR TEIYNWLSSI 540 PSEMESYIRP GCVVLSVYVA MSPAAWEQLE QNLQQRVAVM LQDSHSGFWR DSRFIVNTGR 600 QLASHKNGRI RCSKSWRTWN SPELISVSPV AVVAGEETSL VVRGRSLTND GISIRCTQMG 660 SYVSMEASGA ACKRAIFDEL NVKCFRVNNT QPGFLGRCFI EVENGFRGDS FPLIIANASI 720 CKELNRLEEE FHPKSQEQAQ TSDHRPTSRE ETLCFLNELG WLFQKNQTTE PLEQSEFSLS 780 RFKFLLVCSV ERDYCAVIRT LLDMLVERNV VNDEPNREAL DMLAEIQLLN RAVKRKSTKM 840 VELLIHYSVN LGSSKKLVFL PNITGPGGIT PLHLAACTSD SDDMVDLLTN DPQEIGLSSW 900 NSLCDATGQT PYRYAAMRNN HTYNSLVARK LADKRNKQVS LNIESEIVVD QLGLSRRSST 960 EMNKSSCASC ATVALKYRRR ASGSHRLFPT PIIHSMLAVA TVCVCVCVFM HAFPIVRQGS 1020 HFSWGGLDYG SI* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 1e-29 | 120 | 204 | 1 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Bna.8352 | 0.0 | leaf| seed | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in vasculature of aerial parts. Expressed in cotyledons and leaf petioles. {ECO:0000269|PubMed:15703061}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
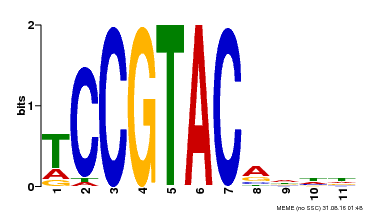
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' (By similarity). May play a role in plant development. {ECO:0000250, ECO:0000269|PubMed:15703061}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00155 | DAP | Transfer from AT1G20980 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | GSBRNA2T00119141001 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK353554 | 0.0 | AK353554.1 Thellungiella halophila mRNA, complete cds, clone: RTFL01-52-I08. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013742040.1 | 0.0 | squamosa promoter-binding-like protein 14 | ||||
| Refseq | XP_013742041.1 | 0.0 | squamosa promoter-binding-like protein 14 | ||||
| Swissprot | Q8RY95 | 0.0 | SPL14_ARATH; Squamosa promoter-binding-like protein 14 | ||||
| TrEMBL | A0A3P6FJQ4 | 0.0 | A0A3P6FJQ4_BRAOL; Uncharacterized protein | ||||
| STRING | Bo5g030930.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3999 | 25 | 54 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G20980.1 | 0.0 | squamosa promoter binding protein-like 14 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



