 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSBRNA2T00120845001 | ||||||||
| Common Name | GSBRNA2T00120845001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 447aa MW: 48982.2 Da PI: 7.5157 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 87.8 | 1.1e-27 | 178 | 230 | 3 | 56 |
G2-like 3 rlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRla 56
+++WtpeLH+rFveaveqL G ekA+P++ilelm+v++Lt+++v+SHLQkYR++
GSBRNA2T00120845001 178 KVDWTPELHRRFVEAVEQL-GLEKAVPSRILELMGVHCLTRHNVASHLQKYRSH 230
789****************.********************************86 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 13.918 | 173 | 232 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.8E-26 | 178 | 233 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.5E-25 | 178 | 230 | IPR006447 | Myb domain, plants |
| SuperFamily | SSF46689 | 3.23E-17 | 179 | 233 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 8.2E-8 | 179 | 228 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0007165 | Biological Process | signal transduction | ||||
| GO:0009658 | Biological Process | chloroplast organization | ||||
| GO:0009910 | Biological Process | negative regulation of flower development | ||||
| GO:0010380 | Biological Process | regulation of chlorophyll biosynthetic process | ||||
| GO:0010638 | Biological Process | positive regulation of organelle organization | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:1900056 | Biological Process | negative regulation of leaf senescence | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 447 aa Download sequence Send to blast |
MLALSPARNS TRDDGESEFL ATSCGFSINP EEDFPEFAVH GDLLDIIDFD DLFGVAGDVL 60 PDLEMDPEIL GGELSDHMNA SSTITTTTSS SEKTKSQGKA NNKKGISGKG EEVVSKRDHN 120 ETPVAETVVN YDGDSGRKRS HSSSGSTKRN PISNNEGKRK VKNHIENKII MFITPFYKVD 180 WTPELHRRFV EAVEQLGLEK AVPSRILELM GVHCLTRHNV ASHLQKYRSH RKHLLAREAE 240 AANWTRKRHI YGLDSTGANA NGRNRNGWLA PAPTLGYPPP PPAAVASPPV HHHHLRPLHV 300 WGHPTVDQSV MPHVWPKHLP PPSTAMATPP FWVSDTPYWP RIHNGTTPYL PTVATRFRTP 360 PVAGIPQALP SHHMVNNYKP DHGFGGPRSL VDLHPSKESV DAAIGDVLTR PWLPLPLGLK 420 PPTVDGVITE LHRHGVSDVP PAASCA* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5lxu_A | 7e-15 | 181 | 232 | 6 | 57 | Transcription factor LUX |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Bna.24050 | 4e-84 | vegetative meristem | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in rosette and cauline leaves. Expressed at low levels in cotyledons and shoots. {ECO:0000269|PubMed:12220263}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that functions with GLK2 to promote chloroplast development. Acts as an activator of nuclear photosynthetic genes involved in chlorophyll biosynthesis, light harvesting, and electron transport. Acts in a cell-autonomous manner to coordinate and maintain the photosynthetic apparatus within individual cells. May function in photosynthetic capacity optimization by integrating responses to variable environmental and endogenous cues (PubMed:11828027, PubMed:12220263, PubMed:17533111, PubMed:18643989, PubMed:19376934, PubMed:19383092, PubMed:19726569). Prevents premature senescence (PubMed:23459204). {ECO:0000269|PubMed:11828027, ECO:0000269|PubMed:12220263, ECO:0000269|PubMed:17533111, ECO:0000269|PubMed:18643989, ECO:0000269|PubMed:19376934, ECO:0000269|PubMed:19383092, ECO:0000269|PubMed:19726569, ECO:0000269|PubMed:23459204}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00022 | PBM | Transfer from AT2G20570 | Download |
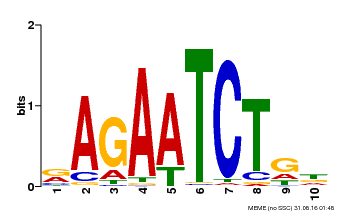
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | GSBRNA2T00120845001 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By light. Repressed by BZR2. {ECO:0000269|PubMed:12220263, ECO:0000269|PubMed:21214652}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013675958.1 | 0.0 | transcription activator GLK1-like isoform X1 | ||||
| Swissprot | Q9SIV3 | 0.0 | GLK1_ARATH; Transcription activator GLK1 | ||||
| TrEMBL | A0A3P6E4A9 | 0.0 | A0A3P6E4A9_BRAOL; Uncharacterized protein | ||||
| STRING | Bo7g003410.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2209 | 27 | 73 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G20570.2 | 1e-173 | GBF's pro-rich region-interacting factor 1 | ||||



