 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSBRNA2T00132911001 | ||||||||
| Common Name | GSBRNA2T00132911001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | Dof | ||||||||
| Protein Properties | Length: 365aa MW: 39921 Da PI: 9.3384 | ||||||||
| Description | Dof family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-Dof | 123.5 | 7.2e-39 | 75 | 135 | 2 | 62 |
zf-Dof 2 kekalkcprCdstntkfCyynnyslsqPryfCkaCrryWtkGGalrnvPvGggrrknkkss 62
++++l+cprC+stntkfCyynny+l+qPryfCk+CrryWt+GG+lrnvPvGg++rknk+ s
GSBRNA2T00132911001 75 PQEKLNCPRCNSTNTKFCYYNNYNLTQPRYFCKGCRRYWTEGGTLRNVPVGGSSRKNKRLS 135
6899******************************************************976 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| ProDom | PD007478 | 2.0E-25 | 74 | 126 | IPR003851 | Zinc finger, Dof-type |
| Pfam | PF02701 | 3.8E-33 | 77 | 133 | IPR003851 | Zinc finger, Dof-type |
| PROSITE profile | PS50884 | 29.031 | 79 | 133 | IPR003851 | Zinc finger, Dof-type |
| PROSITE pattern | PS01361 | 0 | 81 | 117 | IPR003851 | Zinc finger, Dof-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0009416 | Biological Process | response to light stimulus | ||||
| GO:0009845 | Biological Process | seed germination | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 365 aa Download sequence Send to blast |
MDATKWTQGF QEMMNVKPMD QIMIPNNNTH QSNTTSSARP STILTPNGVS AAGATVSGVS 60 NNNNTAVVVE RKARPQEKLN CPRCNSTNTK FCYYNNYNLT QPRYFCKGCR RYWTEGGTLR 120 NVPVGGSSRK NKRLSSSSSS SNILQTTPSS LGLTTTLPDL NPPILFSNQI PNKSKGSSQD 180 LNLLSFPVMQ DQHHRVHMSQ FLQMPKMEGN GNITHQQPSS SSSLYGSSPV SALELLRRGV 240 SVSPNSVMPS GPMMDSSNNV MYTSSGFPTM VDYKPSNLSF STDHHQGTGH NNHNRSDDAH 300 HQGRVLFPFG DQMKELSSSI TQEVDHDDNQ QQKSHGNNNN NISSSPNNGY WSGMFSTTGG 360 GSSW* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Bna.14494 | 0.0 | seed | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Turned off in siliques when they reached full maturation. Not expressed in developing or mature embryos. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in the vascular system of the mother plant, but not present in the seed and embryo. In maturing siliques, found all through the funiculus connecting the placenta to the ovule, but not in the ovule. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor specifically involved in the maternal control of seed germination. Regulates transcription by binding to a 5'-AA[AG]G-3' consensus core sequence. May ensure the activation of a component that would trigger germination as a consequence of red light perception. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00319 | DAP | Transfer from AT2G46590 | Download |
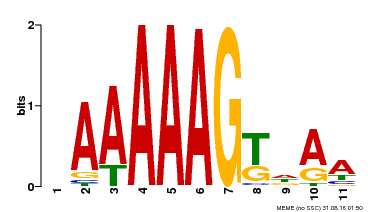
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | GSBRNA2T00132911001 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK352859 | 0.0 | AK352859.1 Thellungiella halophila mRNA, complete cds, clone: RTFL01-17-N20. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009142625.1 | 0.0 | PREDICTED: dof zinc finger protein DOF2.5-like | ||||
| Refseq | XP_013747624.1 | 0.0 | dof zinc finger protein DOF2.5-like | ||||
| Swissprot | Q9ZPY0 | 0.0 | DOF25_ARATH; Dof zinc finger protein DOF2.5 | ||||
| TrEMBL | A0A3P5YNV7 | 0.0 | A0A3P5YNV7_BRACM; Uncharacterized protein | ||||
| STRING | Bra004525.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2528 | 26 | 73 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G46590.2 | 1e-156 | Dof family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



