 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSBRNA2T00135793001 | ||||||||
| Common Name | GSBRNA2T00135793001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 914aa MW: 102262 Da PI: 5.801 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 56.5 | 6.6e-18 | 42 | 88 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg WT+eEde+l +av+ + g++Wk+Ia+ + Rt+ qc +rwqk+l
GSBRNA2T00135793001 42 RGQWTAEEDEILTKAVHSFKGKNWKKIAEFFK-DRTDVQCLHRWQKVL 88
89*****************************9.************986 PP
| |||||||
| 2 | Myb_DNA-binding | 62.9 | 6.5e-20 | 94 | 140 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEde +v++++++G++ W+tIar ++ gR +kqc++rw+++l
GSBRNA2T00135793001 94 KGPWTKEEDEMIVQLIQKYGPKKWSTIARFLP-GRIGKQCRERWHNHL 140
79******************************.*************97 PP
| |||||||
| 3 | Myb_DNA-binding | 43.1 | 9.8e-14 | 146 | 188 | 1 | 45 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwq 45
+ +WT+eE++ l++a++ +G++ W+ + ++ gR+++ +k++w+
GSBRNA2T00135793001 146 KEAWTQEEELVLIRAHQIYGNR-WAELTKFLP-GRSDNGIKNHWH 188
579*******************.*********.***********8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 17.673 | 37 | 88 | IPR017930 | Myb domain |
| SMART | SM00717 | 4.6E-15 | 41 | 90 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.6E-15 | 42 | 88 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 2.58E-15 | 43 | 98 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.4E-22 | 43 | 96 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.77E-13 | 45 | 88 | No hit | No description |
| PROSITE profile | PS51294 | 32.297 | 89 | 144 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.12E-31 | 91 | 187 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 5.1E-19 | 93 | 142 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.0E-17 | 94 | 140 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 8.91E-17 | 96 | 140 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 2.0E-27 | 97 | 144 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.5E-12 | 145 | 193 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 18.094 | 145 | 195 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.5E-19 | 145 | 195 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 9.0E-12 | 146 | 188 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 9.42E-10 | 148 | 188 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0032875 | Biological Process | regulation of DNA endoreduplication | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003713 | Molecular Function | transcription coactivator activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 914 aa Download sequence Send to blast |
MESECTTTTS STPPPPPPQG TTQGNPKSRQ GRTSGPARRS TRGQWTAEED EILTKAVHSF 60 KGKNWKKIAE FFKDRTDVQC LHRWQKVLNP ELVKGPWTKE EDEMIVQLIQ KYGPKKWSTI 120 ARFLPGRIGK QCRERWHNHL NPAINKEAWT QEEELVLIRA HQIYGNRWAE LTKFLPGRSD 180 NGIKNHWHSS VKKKLDSYMS SGLLDQYQAM PLAPYDRNSA LQSHFMQSTM DGSGCLSGQA 240 EQEIENCQIS SVADQNGLMN MGQHFHPCEN SQYYYPELED ISVSISETSY DMEDCSQFPD 300 HNVSASTSQD YHFDFQDISL EMSHDMSELP IPYTKERKEA SLGAPNSTSN IDVAAYTNTS 360 ETECCRVLFL DQESEGVSVS RSSTEEPHEG LCASASDSQV SEATKSPMKS SSSTSIATPA 420 SGKETLRPSP LIITPDKYSK KSSGLICHPF EAEPNCRTDE NGSFICINDP SNSTCVDEGP 480 SYHLNDSKKL VPVNDFTSLA DVKPQPSLPK HETNMSREQH HEDMGASSSS LCFPSLDLPA 540 FNDPVHDYSP LGIRKLLMST MTCMSPLRLW ESPTGKKTLV GAKSILRKRT RDLLTPLSEK 600 RSDKKLETDI AASLAKDFSR LDVMFDESEN QESISGTSLV SHRMPEETTD VSKSLEKQQQ 660 TCLEANVQHA ESFSGVLSES NTNKQVLSPP GQSLTKAEKT QVSTPRNHLQ RTLMATSNKE 720 QHSSPSLCLV INSPSRARNT EGHLANNETN NENFSIFCGT PFRRGLESPS AWKSPFYVNS 780 LLPSPRFDTD ITIEDMGYIF SPGERSYESR GMFTQRHEHT SAFAAFNAME ISLSPSTDDA 840 KKMKDLDKEN NDPLMAEGRV LDFNDCESPT NHPRLSKQAD SRSDINSLVP IHGWGLLSFY 900 MHGERFWIIT SYE* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h88_C | 9e-68 | 42 | 195 | 6 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 9e-68 | 42 | 195 | 6 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| Search in ModeBase | ||||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Expressed in the root quiescent center (PubMed:15937229). Accumulates in division zone of primary root tips and emerging lateral roots. Also present in floral organs in young flower buds. Weakly and uniformly present in the developing embryo and maternal tissues (PubMed:17287251). Expressed specifically in proliferating stage of leaves (PubMed:26069325). {ECO:0000269|PubMed:15937229, ECO:0000269|PubMed:17287251, ECO:0000269|PubMed:26069325}. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in roots, cotyledons and leaves, especially in vascular tissues, and in flowers. {ECO:0000269|PubMed:17287251}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds 5'-AACGG-3' motifs in gene promoters (PubMed:21862669). Involved in the regulation of cytokinesis, probably via the activation of several G2/M phase-specific genes transcription (e.g. KNOLLE) (PubMed:17287251, PubMed:21862669, PubMed:25806785). Required for the maintenance of diploidy (PubMed:21862669). {ECO:0000269|PubMed:17287251, ECO:0000269|PubMed:21862669, ECO:0000269|PubMed:25806785}.; FUNCTION: Involved in transcription regulation during induced endoreduplication at the powdery mildew (e.g. G.orontii) infection site, thus promoting G.orontii growth and reproduction. {ECO:0000269|PubMed:20018666}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00505 | DAP | Transfer from AT5G11510 | Download |
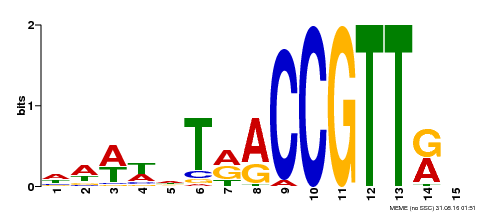
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | GSBRNA2T00135793001 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Slightly induced by salicylic acid (SA) (PubMed:16463103). Expressed in a cell cycle-dependent manner, with highest levels 2 hours before the peak of mitotic index in cells synchronized by aphidicolin. Activated by CYCB1 (PubMed:17287251). Accumulates at powdery mildew (e.g. G.orontii) infected cells. {ECO:0000269|PubMed:16463103, ECO:0000269|PubMed:17287251}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AF371975 | 0.0 | AF371975.2 Arabidopsis thaliana putative c-myb-like transcription factor MYB3R-4 (MYB3R4) mRNA, complete cds. | |||
| GenBank | AY519650 | 0.0 | AY519650.1 Arabidopsis thaliana MYB transcription factor (At5g11510) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013607014.1 | 0.0 | PREDICTED: LOW QUALITY PROTEIN: myb-related protein 3R-1 | ||||
| Refseq | XP_022564903.1 | 0.0 | LOW QUALITY PROTEIN: transcription factor MYB3R-4-like | ||||
| Swissprot | Q94FL9 | 0.0 | MB3R4_ARATH; Transcription factor MYB3R-4 | ||||
| TrEMBL | A0A3P6D240 | 0.0 | A0A3P6D240_BRACM; Uncharacterized protein | ||||
| STRING | Bra008956.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM13213 | 18 | 28 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G11510.1 | 0.0 | myb domain protein 3r-4 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



