 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSBRNA2T00146726001 | ||||||||
| Common Name | GSBRNA2T00146726001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 295aa MW: 33884.9 Da PI: 6.6772 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 162.8 | 1.2e-50 | 6 | 134 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLp.k.kvkaeekewyfFskrdkkyatgkrknratksgyWkatgk 88
lppGfrFhPtdeel+++yLk+kveg ++el evi+ +d+yk++ w+Lp k + ++ ew+fF++rdkky++g r+nr tk+gyWkatgk
GSBRNA2T00146726001 6 LPPGFRFHPTDEELIDYYLKRKVEGLEIEL-EVIPVIDLYKFDHWELPdKsFLPNRGMEWFFFCSRDKKYPNGFRTNRGTKAGYWKATGK 94
79****************************.99**************95434555778******************************** PP
NAM 89 dkevlskkgelvglkktLvfykgrapkgektdWvmheyrl 128
d++++s+++++vg +ktLvfykgrap g +t+W+mheyrl
GSBRNA2T00146726001 95 DRKITSRSSSTVGYRKTLVFYKGRAPLGDRTTWLMHEYRL 134
**************************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 3.66E-57 | 3 | 159 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 56.65 | 6 | 159 | IPR003441 | NAC domain |
| Pfam | PF02365 | 4.8E-28 | 7 | 134 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 295 aa Download sequence Send to blast |
MGSSCLPPGF RFHPTDEELI DYYLKRKVEG LEIELEVIPV IDLYKFDHWE LPDKSFLPNR 60 GMEWFFFCSR DKKYPNGFRT NRGTKAGYWK ATGKDRKITS RSSSTVGYRK TLVFYKGRAP 120 LGDRTTWLMH EYRLRDDESS SQGSQTYKGA YVLCRVAKKN EIKTNSKIRR NLSEQTLGSG 180 EISGYSSRVT SPSRDGTMPF HSFVNPVSTE IDSSNIWISP DFILDSSKDY PQIQNFASEY 240 FQDFDFPVIG QEVNFPASIL HTDVDQNMDE SMQTGYWTNC GYNQISLFGY SELS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 4e-49 | 6 | 160 | 17 | 166 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 4e-49 | 6 | 160 | 17 | 166 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 4e-49 | 6 | 160 | 17 | 166 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 4e-49 | 6 | 160 | 17 | 166 | NO APICAL MERISTEM PROTEIN |
| 3swm_A | 5e-49 | 6 | 160 | 20 | 169 | NAC domain-containing protein 19 |
| 3swm_B | 5e-49 | 6 | 160 | 20 | 169 | NAC domain-containing protein 19 |
| 3swm_C | 5e-49 | 6 | 160 | 20 | 169 | NAC domain-containing protein 19 |
| 3swm_D | 5e-49 | 6 | 160 | 20 | 169 | NAC domain-containing protein 19 |
| 3swp_A | 5e-49 | 6 | 160 | 20 | 169 | NAC domain-containing protein 19 |
| 3swp_B | 5e-49 | 6 | 160 | 20 | 169 | NAC domain-containing protein 19 |
| 3swp_C | 5e-49 | 6 | 160 | 20 | 169 | NAC domain-containing protein 19 |
| 3swp_D | 5e-49 | 6 | 160 | 20 | 169 | NAC domain-containing protein 19 |
| 4dul_A | 4e-49 | 6 | 160 | 17 | 166 | NAC domain-containing protein 19 |
| 4dul_B | 4e-49 | 6 | 160 | 17 | 166 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in roots, rosettes leaves, cauline leaves and stems. {ECO:0000269|PubMed:24285786}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator involved in the positive regulation of abscisic acid (ABA) responsive genes. Acts as a positive factor of ABA-mediated responses. Involved in the transcriptional activation of ABA-inducible genes in response to dehydration and osmotic stresses. Plays a positive role in both stomatal closure and water loss under dehydration stress conditions. Acts synergistically with ABF2 to activate the dehydration stress-response factor RD29A transcription. Binds to the consensus core cis-acting elements 5'-CGTA-3' and 5'-CACG-3' at the RD29A promoter (PubMed:24285786). Involved in hypocotyl graft union formation. Required for the auxin-mediated promotion of vascular tissue proliferation during hypocotyl graft attachment (PubMed:25182467). {ECO:0000269|PubMed:24285786, ECO:0000269|PubMed:25182467}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00546 | DAP | Transfer from AT5G46590 | Download |
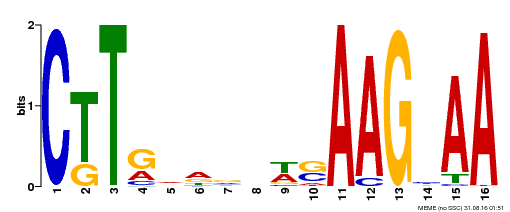
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | GSBRNA2T00146726001 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by abscisic acid (ABA), dehydration and osmotic stress. {ECO:0000269|PubMed:24285786}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009128881.1 | 0.0 | PREDICTED: NAC domain-containing protein 86-like | ||||
| Swissprot | Q9LS24 | 1e-171 | NAC96_ARATH; NAC domain-containing protein 96 | ||||
| TrEMBL | A0A3P6E1T8 | 0.0 | A0A3P6E1T8_BRAOL; Uncharacterized protein | ||||
| STRING | Bra022046.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2151 | 27 | 77 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G46590.1 | 1e-173 | NAC domain containing protein 96 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



