 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSBRNA2T00150520001 | ||||||||
| Common Name | GSBRNA2T00150520001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 541aa MW: 59461.7 Da PI: 6.5773 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 39.5 | 1e-12 | 382 | 427 | 4 | 54 |
HHHHHHHHHHHHHHHHHHHHHCTSCC.C...TTS-STCHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPk.askapskKlsKaeiLekAveYIksL 54
+h e+Er+RR+++N++f Lr+++P+ + K++Ka+ L Av YI++L
GSBRNA2T00150520001 382 NHVEAERQRREKLNQRFYALRSVVPNiS------KMDKASLLGDAVSYINEL 427
799***********************66......***************988 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 5.3E-49 | 9 | 199 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| PROSITE profile | PS50888 | 17.326 | 378 | 427 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.83E-18 | 378 | 447 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 5.79E-14 | 381 | 432 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 9.0E-18 | 382 | 441 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 2.7E-10 | 382 | 427 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 5.6E-16 | 384 | 433 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 541 aa Download sequence Send to blast |
MTVGSDDNLQ NKLSDLVETP NSSNFSWNYA IFWQVSRSKS GDLVLCWGDG SCREPKDGER 60 SEMMRMLSMG REEETHQTLR KRVLQKLHAL FGGLDDEEDS CALVLDRVTD TEMFLLASMY 120 FSFPRGQGGP GKCFHSSQPV WLSDLVNSGS DYCVRSFLAK SAGIQTVVLV PTDIGVVELG 180 STSCLPHSDE SLSSIRLSFS SPPPPPPPPP PVRLPVVVAN HNDDNSSSKI FGKDLHSSSG 240 FLHQQQHRQF REKLTVRKMD DNRVPKRLDG NNNRFMFSNP ITTMTTSTWI PPEAPTREDF 300 KFLPLQQTSS SSHRLLPPAQ MQIDFSGASS RPSDNNSDGG GGGGGGGADW SDLVGGVDES 360 GGDNRPRKRG RRPANGRAEA LNHVEAERQR REKLNQRFYA LRSVVPNISK MDKASLLGDA 420 VSYINELYAK LKVMEAEREK LGYSSNPLIC LEPPAVNVQT AGEDVAVTVN CSLDSHPASR 480 IFHAFEEAKV EVINAKLEVP QDVVLHTFVI KSEEVTKEKL ISALSREQSG SVQSRTSSGR 540 * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5gnj_A | 2e-26 | 376 | 441 | 2 | 65 | Transcription factor MYC2 |
| 5gnj_B | 2e-26 | 376 | 441 | 2 | 65 | Transcription factor MYC2 |
| 5gnj_E | 2e-26 | 376 | 441 | 2 | 65 | Transcription factor MYC2 |
| 5gnj_F | 2e-26 | 376 | 441 | 2 | 65 | Transcription factor MYC2 |
| 5gnj_G | 2e-26 | 376 | 441 | 2 | 65 | Transcription factor MYC2 |
| 5gnj_I | 2e-26 | 376 | 441 | 2 | 65 | Transcription factor MYC2 |
| 5gnj_M | 2e-26 | 376 | 441 | 2 | 65 | Transcription factor MYC2 |
| 5gnj_N | 2e-26 | 376 | 441 | 2 | 65 | Transcription factor MYC2 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Bna.13201 | 0.0 | microspore | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00100 | PBM | Transfer from AT1G01260 | Download |
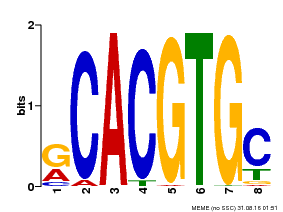
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | GSBRNA2T00150520001 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By UV treatment. {ECO:0000269|PubMed:12679534}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_022548937.1 | 0.0 | transcription factor bHLH13-like isoform X1 | ||||
| Refseq | XP_022548938.1 | 0.0 | transcription factor bHLH13-like isoform X2 | ||||
| Swissprot | Q9LNJ5 | 0.0 | BH013_ARATH; Transcription factor bHLH13 | ||||
| TrEMBL | M4EWT8 | 0.0 | M4EWT8_BRARP; Uncharacterized protein | ||||
| STRING | Bra033273.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3758 | 27 | 59 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G01260.3 | 0.0 | bHLH family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



