 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSMUA_Achr10P21480_001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Zingiberales; Musaceae; Musa
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 319aa MW: 36829.2 Da PI: 8.9369 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 98 | 3.9e-31 | 86 | 136 | 1 | 51 |
S---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE- CS
SRF-TF 1 krienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklyeyss 51
krien++nrqvtf+kRrng+lKKA+ELSvLCda+va+i+fss+g+lyey++
GSMUA_Achr10P21480_001 86 KRIENTTNRQVTFCKRRNGLLKKAYELSVLCDADVALIVFSSRGRLYEYAT 136
79***********************************************86 PP
| |||||||
| 2 | K-box | 111 | 1.3e-36 | 157 | 250 | 8 | 100 |
K-box 8 s.leeakaeslqqelakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkekelqeenka 93
++e +a+++qqe++kL+++i+nLq+++R l+Ge+L+s+sl++++qLe++Lek+++kiR+kKnell+++ie++qk+e elq++n++
GSMUA_Achr10P21480_001 157 GfASEDNAQYYQQEASKLRQQINNLQSTNRSLMGESLGSMSLRDMKQLETRLEKGINKIRNKKNELLFAEIEYMQKREMELQNDNMY 243
4578999******************************************************************************** PP
K-box 94 Lrkklee 100
Lr+k++e
GSMUA_Achr10P21480_001 244 LRNKIAE 250
****986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00432 | 6.6E-43 | 78 | 137 | IPR002100 | Transcription factor, MADS-box |
| PROSITE profile | PS50066 | 34.048 | 78 | 138 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 1.18E-45 | 79 | 152 | No hit | No description |
| SuperFamily | SSF55455 | 4.19E-33 | 79 | 152 | IPR002100 | Transcription factor, MADS-box |
| PROSITE pattern | PS00350 | 0 | 80 | 134 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 2.6E-32 | 80 | 100 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 1.1E-26 | 87 | 134 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 2.6E-32 | 100 | 115 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 2.6E-32 | 115 | 136 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF01486 | 4.5E-28 | 161 | 248 | IPR002487 | Transcription factor, K-box |
| PROSITE profile | PS51297 | 15.59 | 164 | 254 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0048440 | Biological Process | carpel development | ||||
| GO:0048443 | Biological Process | stamen development | ||||
| GO:0048497 | Biological Process | maintenance of floral organ identity | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 319 aa Download sequence Send to blast |
MDEDFTLFEC CHDSTLIQKY FPLTIINVPL NTQERGIAEL LVTIAVPFSP QTFFFASASA 60 FLLLLLRRGA TLEPPARMGR GKIEIKRIEN TTNRQVTFCK RRNGLLKKAY ELSVLCDADV 120 ALIVFSSRGR LYEYATNSVK ATIDRYKKAC NGTTNTGFAS EDNAQYYQQE ASKLRQQINN 180 LQSTNRSLMG ESLGSMSLRD MKQLETRLEK GINKIRNKKN ELLFAEIEYM QKREMELQND 240 NMYLRNKIAE NERAQQQMSM LPSARTTEYE IMPPYDSRNF LQVNAVQPTQ HYTHQQRTAL 300 QLGTPCTRWV MCSCCHFCR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1tqe_P | 1e-19 | 78 | 146 | 1 | 69 | Myocyte-specific enhancer factor 2B |
| 1tqe_Q | 1e-19 | 78 | 146 | 1 | 69 | Myocyte-specific enhancer factor 2B |
| 1tqe_R | 1e-19 | 78 | 146 | 1 | 69 | Myocyte-specific enhancer factor 2B |
| 1tqe_S | 1e-19 | 78 | 146 | 1 | 69 | Myocyte-specific enhancer factor 2B |
| 6c9l_A | 1e-19 | 78 | 146 | 1 | 69 | Myocyte-specific enhancer factor 2B |
| 6c9l_B | 1e-19 | 78 | 146 | 1 | 69 | Myocyte-specific enhancer factor 2B |
| 6c9l_C | 1e-19 | 78 | 146 | 1 | 69 | Myocyte-specific enhancer factor 2B |
| 6c9l_D | 1e-19 | 78 | 146 | 1 | 69 | Myocyte-specific enhancer factor 2B |
| 6c9l_E | 1e-19 | 78 | 146 | 1 | 69 | Myocyte-specific enhancer factor 2B |
| 6c9l_F | 1e-19 | 78 | 146 | 1 | 69 | Myocyte-specific enhancer factor 2B |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in regulating genes that determines stamen and carpel development in wild-type flowers. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00609 | ChIP-seq | Transfer from AT4G18960 | Download |
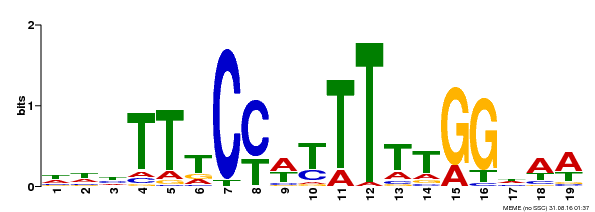
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018674455.1 | 1e-166 | PREDICTED: MADS-box transcription factor 3-like isoform X2 | ||||
| Swissprot | Q40872 | 1e-119 | AG_PANGI; Floral homeotic protein AGAMOUS | ||||
| TrEMBL | M0RK73 | 0.0 | M0RK73_MUSAM; Uncharacterized protein | ||||
| STRING | GSMUA_Achr10P21480_001 | 0.0 | (Musa acuminata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP649 | 37 | 144 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G18960.1 | 8e-99 | MIKC_MADS family protein | ||||



