 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSMUA_Achr10P23260_001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Zingiberales; Musaceae; Musa
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 469aa MW: 52123.6 Da PI: 6.5109 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 101.6 | 5e-32 | 260 | 314 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
k+rlrWt eLHerFveav++L G+ekAtPk +l+lm+v+gLt++hvkSHLQkYRl
GSMUA_Achr10P23260_001 260 KTRLRWTLELHERFVEAVNKLDGAEKATPKGVLKLMNVEGLTIYHVKSHLQKYRL 314
68****************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 10.544 | 257 | 317 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.2E-30 | 258 | 315 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 3.23E-17 | 259 | 314 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 4.2E-24 | 260 | 315 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 3.0E-8 | 262 | 313 | IPR001005 | SANT/Myb domain |
| Pfam | PF14379 | 3.0E-22 | 351 | 398 | IPR025756 | MYB-CC type transcription factor, LHEQLE-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 469 aa Download sequence Send to blast |
MSSHSLITVK QNNSPEGTKH LCHSSPLSVQ FNNEQDCQKL SRSSYVETVL HNSSSFSKDL 60 PLHTKKSSSG PEPGSSCSYV SHPQYPEHMF SRSSTFCTSL YSSSSTSSES CQKLQNLPFL 120 PHPPKCKHQN SAVQSSNSPL QFSGDISATS GEDEHTDDLM KDFLNLSGEV SDGSIHGENC 180 GSSGLALNEQ IELQILSEQL GIAITDNGES PHLDDIYETP QVSSLPLSAN HNQTDQPSKP 240 HTKVQLHSPP SINPASAANK TRLRWTLELH ERFVEAVNKL DGAEKATPKG VLKLMNVEGL 300 TIYHVKSHLQ KYRLAKYLPE AKEDKKASSR EDKKSSSLVS NDGDLANKRS IQVTEALRMQ 360 IEVQKQLHEQ LEVQRALQLR IEENARYLQK ILEEQQKANN SSSSTQRFSS EPPLELRSPF 420 TEKADARVDS SPLNSVKQRG NESNDRESDS ESVEDSKRMR LDVEHIRPS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 7e-25 | 260 | 317 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 7e-25 | 260 | 317 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 7e-25 | 260 | 317 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 7e-25 | 260 | 317 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 7e-25 | 260 | 317 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 7e-25 | 260 | 317 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 7e-25 | 260 | 317 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 7e-25 | 260 | 317 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in phosphate starvation signaling (PubMed:26082401). Binds to P1BS, an imperfect palindromic sequence 5'-GNATATNC-3', to promote the expression of inorganic phosphate (Pi) starvation-responsive genes (PubMed:26082401). Functionally redundant with PHR1 and PHR2 in regulating Pi starvation response and Pi homeostasis (PubMed:26082401). {ECO:0000269|PubMed:26082401}. | |||||
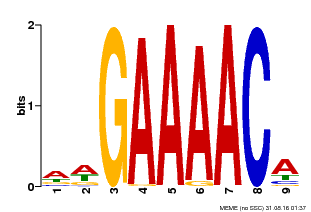
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00354 | DAP | Transfer from AT3G13040 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated under Pi starvation conditions. {ECO:0000269|PubMed:26082401}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018674430.1 | 0.0 | PREDICTED: protein PHOSPHATE STARVATION RESPONSE 3-like | ||||
| Refseq | XP_018674431.1 | 0.0 | PREDICTED: protein PHOSPHATE STARVATION RESPONSE 3-like | ||||
| Refseq | XP_018674432.1 | 0.0 | PREDICTED: protein PHOSPHATE STARVATION RESPONSE 3-like | ||||
| Swissprot | Q6YXZ4 | 1e-120 | PHR3_ORYSJ; Protein PHOSPHATE STARVATION RESPONSE 3 | ||||
| TrEMBL | M0RKQ1 | 0.0 | M0RKQ1_MUSAM; Uncharacterized protein | ||||
| STRING | GSMUA_Achr10P23260_001 | 0.0 | (Musa acuminata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP4526 | 37 | 69 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G13040.2 | 1e-93 | G2-like family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



