 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSMUA_Achr11P02230_001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Zingiberales; Musaceae; Musa
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 340aa MW: 37421.5 Da PI: 8.7152 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 21.9 | 4.6e-07 | 191 | 213 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
+ C+ Cgk F+r nL+ H+r H
GSMUA_Achr11P02230_001 191 HYCQVCGKGFRRDANLRMHMRAH 213
68*******************99 PP
| |||||||
| 2 | zf-C2H2 | 11.7 | 0.00077 | 279 | 302 | 1 | 23 |
EEETTTT.EEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCg.ksFsrksnLkrHirtH 23
y+C+ C+ k Fs s+L++H + +
GSMUA_Achr11P02230_001 279 YVCNRCNlKQFSVLSDLRTHEKHC 302
9*******************9876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF57667 | 3.31E-6 | 189 | 216 | No hit | No description |
| SMART | SM00355 | 0.0039 | 191 | 213 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 12.715 | 191 | 218 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 9.9E-7 | 191 | 217 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 193 | 213 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 150 | 241 | 274 | IPR015880 | Zinc finger, C2H2-like |
| SMART | SM00355 | 24 | 279 | 301 | IPR015880 | Zinc finger, C2H2-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 340 aa Download sequence Send to blast |
MDDILYSSAG AAASGTDQIT SSHALLYSLS VLREKVQQLE SCVSMMTSPN RTQQESIAMG 60 VSCAGIMIQE IILAASSLSC TLPQVGPSTA VAYEEPPPSD MKVESATAWM DHSSNNSLGI 120 TRKDDFFSSN NPSANSAIDV DNGSVTRDQI RKVKPPLDRA GMRKECSQGL SSNGYTIIEL 180 DAADLLAKYT HYCQVCGKGF RRDANLRMHM RAHGDEYKSA AALANPTKSS RSSSSGVALK 240 YSCPHDGCRW NRKHAKFQPL KSVVCAKNHY KRSHCPKMYV CNRCNLKQFS VLSDLRTHEK 300 HCGDLRWRCS CGTNFSRKDK LMGHVALFVG HTPEPCRLRD |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. Together with STOP1, plays a critical role in tolerance to major stress factors in acid soils such as proton H(+) and aluminum ion Al(3+). Required for the expression of genes in response to acidic stress (e.g. ALMT1 and MATE), and Al-activated citrate exudation. {ECO:0000269|PubMed:23935008}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00522 | DAP | Transfer from AT5G22890 | Download |
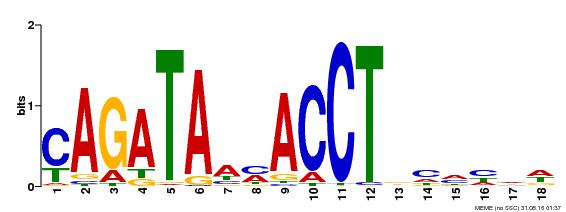
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009382160.1 | 0.0 | PREDICTED: zinc finger protein STAR3-like | ||||
| Swissprot | Q0WT24 | 1e-101 | STOP2_ARATH; Protein SENSITIVE TO PROTON RHIZOTOXICITY 2 | ||||
| TrEMBL | M0RNQ3 | 0.0 | M0RNQ3_MUSAM; Uncharacterized protein | ||||
| STRING | GSMUA_Achr11P02230_001 | 0.0 | (Musa acuminata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP7441 | 32 | 51 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G22890.1 | 1e-103 | C2H2 family protein | ||||



