 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSMUA_Achr11P03290_001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Zingiberales; Musaceae; Musa
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 776aa MW: 83705.2 Da PI: 6.4642 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 66.6 | 3.2e-21 | 114 | 169 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
r++ +++t++q++eLe+lF+++++p+ ++r eL+++l L+ rqVk+WFqNrR+++k
GSMUA_Achr11P03290_001 114 RKRYHRHTPQQIQELEALFKECPHPDDKQRMELSNRLCLEARQVKFWFQNRRTQMK 169
789999***********************************************998 PP
| |||||||
| 2 | START | 182.3 | 2.7e-57 | 302 | 524 | 1 | 203 |
HHHHHHHHHHHHHHHHC-TT-EEEE....EXCCTTEEEEEEESSS......SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S. CS
START 1 elaeeaaqelvkkalaeepgWvkss....esengdevlqkfeeskv.....dsgealrasgvvdmvlallveellddkeqWdetla. 77
ela++a++elvk+a+ eep+W + +n+de+++ f+ + + +ea r+++v +++ lve+l+d + +W +++
GSMUA_Achr11P03290_001 302 ELALAAMDELVKMAQMEEPLWIPGLeagsDALNYDEYYRCFSGCIGarptgFVSEATRETAVIVINSPALVETLMDAA-RWADMFPs 387
5899*****************98888899************66555999999**********9999999*********.******** PP
...EEEEEEEECTT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--..-TTSEE-EESSEEEE CS
START 78 ...kaetlevissg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppe.sssvvRaellpSgil 153
+ +t++vissg galqlm+aelq+lsplvp Rd+ f+R+++ l++g w++vdvSvd +++ + v++++lpSg++
GSMUA_Achr11P03290_001 388 viaRTTTTDVISSGmggtrnGALQLMQAELQVLSPLVPvRDVSFLRFCKHLSEGAWAVVDVSVDGVRDNRPaPPATVKCRRLPSGCV 474
*99*************************************************************99999888999************ PP
EEEECTCEEEEEEEE-EE--SSXXHHHHHHHHHHHHHHHHHHHHHHTXXX CS
START 154 iepksnghskvtwvehvdlkgrlphwllrslvksglaegaktwvatlqrq 203
++++sng+skvtwveh +++++++h l+r+l++sg a ga++wva lqrq
GSMUA_Achr11P03290_001 475 VQDMSNGYSKVTWVEHSEYDEATVHPLYRPLLRSGSALGARRWVASLQRQ 524
************************************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 5.13E-20 | 102 | 169 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 8.9E-22 | 103 | 169 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 16.827 | 111 | 171 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 7.9E-15 | 112 | 175 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 4.31E-18 | 113 | 169 | No hit | No description |
| Pfam | PF00046 | 8.8E-19 | 114 | 169 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 146 | 169 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 41.315 | 293 | 530 | IPR002913 | START domain |
| CDD | cd08875 | 2.57E-115 | 299 | 525 | No hit | No description |
| SuperFamily | SSF55961 | 1.26E-30 | 300 | 525 | No hit | No description |
| Pfam | PF01852 | 2.8E-49 | 302 | 524 | IPR002913 | START domain |
| SMART | SM00234 | 5.1E-40 | 302 | 527 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 1.56E-20 | 544 | 742 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009827 | Biological Process | plant-type cell wall modification | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0043481 | Biological Process | anthocyanin accumulation in tissues in response to UV light | ||||
| GO:0048765 | Biological Process | root hair cell differentiation | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 776 aa Download sequence Send to blast |
MTFGGLFDDG SGGGVARLVA DIAYGGTADS VSHPRRLVSP SLHESMFASP GLSLALQTNL 60 GAHGERNLAR VGGVGGELDS VRRSKEDENE SRAGSDNFEG GSGDDLEQEH PRKRKRYHRH 120 TPQQIQELEA LFKECPHPDD KQRMELSNRL CLEARQVKFW FQNRRTQMKV FTHLERHENT 180 ILRQENDKLR AENLSIRDAM RNPICCNCGS PAVLGEISLE EQHLRIENAR LKDELDRVCA 240 LAGKFLGKPA SPLASPLPLP MLPNSSLELA VGTNGFAGLA QVVGAGATGA VDKAQERFVF 300 LELALAAMDE LVKMAQMEEP LWIPGLEAGS DALNYDEYYR CFSGCIGARP TGFVSEATRE 360 TAVIVINSPA LVETLMDAAR WADMFPSVIA RTTTTDVISS GMGGTRNGAL QLMQAELQVL 420 SPLVPVRDVS FLRFCKHLSE GAWAVVDVSV DGVRDNRPAP PATVKCRRLP SGCVVQDMSN 480 GYSKVTWVEH SEYDEATVHP LYRPLLRSGS ALGARRWVAS LQRQSLAVLV PPSLSPGGDS 540 TITPSGRRSM LKLAQRMTDN FCAGVCASSA REWSKLGGAI NIGEDVRVMT RQSVADPGVP 600 PGVVLSAATS VWLPASPQRL FDFLRNEQLR SQWDILSNGG PMQEIAHIAK GQNTGNAVSL 660 LRASAANATQ SSMLILQETC TDASGSLVVY APVDIPAMHL VMSGGDSAYV SLLPSGFAIL 720 PDGGTHKAPG SLLTVAFQIL VNSQPTAKLT VESVETVNNL ISCTVQKIKA ALHCET |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of the tissue-specific accumulation of anthocyanins and in cellular organization of the primary root. {ECO:0000269|PubMed:10402424}. | |||||
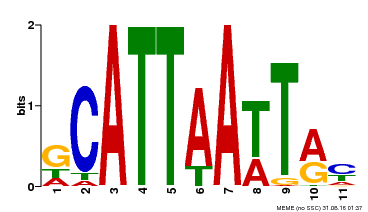
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00421 | DAP | Transfer from AT4G00730 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018676535.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein ROC5-like | ||||
| Refseq | XP_018676536.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein ROC5-like | ||||
| Swissprot | Q0WV12 | 0.0 | ANL2_ARATH; Homeobox-leucine zipper protein ANTHOCYANINLESS 2 | ||||
| TrEMBL | M0RP09 | 0.0 | M0RP09_MUSAM; Uncharacterized protein | ||||
| STRING | GSMUA_Achr11P03290_001 | 0.0 | (Musa acuminata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1203 | 37 | 116 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00730.1 | 0.0 | HD-ZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



