 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSMUA_Achr11P06340_001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Zingiberales; Musaceae; Musa
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 283aa MW: 31610.7 Da PI: 9.0948 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 105.9 | 2.3e-33 | 33 | 87 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
kprlrWtp+LH+rFv+av++LGG++kAtPk++l lm++kgLtl+h+kSHLQkYRl
GSMUA_Achr11P06340_001 33 KPRLRWTPDLHDRFVDAVTKLGGPDKATPKSVLSLMGMKGLTLYHLKSHLQKYRL 87
79****************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 4.6E-32 | 30 | 88 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 12.757 | 30 | 90 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 5.2E-16 | 33 | 88 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 8.2E-25 | 33 | 88 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 3.1E-9 | 35 | 86 | IPR001005 | SANT/Myb domain |
| Pfam | PF14379 | 2.9E-22 | 129 | 175 | IPR025756 | MYB-CC type transcription factor, LHEQLE-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 283 aa Download sequence Send to blast |
MENIYQRRPP LAASLFEAPG GEGAGVMLSR DPKPRLRWTP DLHDRFVDAV TKLGGPDKAT 60 PKSVLSLMGM KGLTLYHLKS HLQKYRLGKQ TRRETEQEAK KKGSNSSKIN CSSTTSNYVS 120 RTDGAGEMPL GEALCYQIEV QRKLQEQLEV QKKLQTRIEA QGKYLQAILE KAQKSLCFDN 180 NRSSGSLEAT RAQLTDFDLP QSGLMENVGR VCEEKHSELR EVRPQENIKK RNNSGFQLYQ 240 EGRDEAEDSS LLLLDLNVKG SSGEMVGGSR GNDLDLRIQT QGL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4r_A | 2e-19 | 33 | 89 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 2e-19 | 33 | 89 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 2e-19 | 33 | 89 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 2e-19 | 33 | 89 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00544 | DAP | Transfer from AT5G45580 | Download |
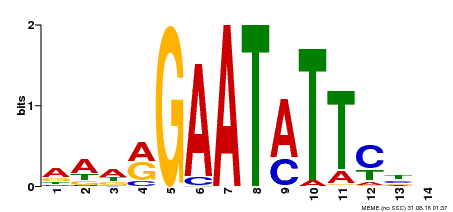
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009382623.1 | 0.0 | PREDICTED: myb family transcription factor PHL11-like | ||||
| Swissprot | C0SVS4 | 2e-65 | PHLB_ARATH; Myb family transcription factor PHL11 | ||||
| TrEMBL | M0RPW4 | 0.0 | M0RPW4_MUSAM; Uncharacterized protein | ||||
| STRING | GSMUA_Achr11P06340_001 | 0.0 | (Musa acuminata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3940 | 34 | 70 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45580.1 | 2e-54 | G2-like family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



