 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSMUA_Achr11P09440_001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Zingiberales; Musaceae; Musa
|
||||||||
| Family | BBR-BPC | ||||||||
| Protein Properties | Length: 270aa MW: 29829.9 Da PI: 10.3838 | ||||||||
| Description | BBR-BPC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GAGA_bind | 31.5 | 5.1e-10 | 1 | 99 | 1 | 104 |
GAGA_bind 1 mdddgsre.rnkgyyepaaslkenlglqlmssiaerdakirernlalsekkaavaerd..maflqrdkalaernkalverdnkllal 84
mdddg rn+gyyep+ k nlgl+lmss e+dak+ +n+ +++++ ++ e ++f ++d ++++ n++ +n ++
GSMUA_Achr11P09440_001 1 MDDDGGLGiRNWGYYEPS--SKGNLGLRLMSSTVEQDAKPLLSNAGFIHRHCSIPEPTapVDF-MSDGWFHHSNDNS---KNDYVRD 81
999999999*******99..799***************************9999765545899.99*****944432...3333333 PP
GAGA_bind 85 llvenslasalpvgvqvlsg 104
l+++s + ++++ ++l+
GSMUA_Achr11P09440_001 82 GLMRHSNN--NSKNLHILPT 99
33333322..2222333333 PP
| |||||||
| 2 | GAGA_bind | 266.4 | 1.6e-81 | 108 | 270 | 136 | 301 |
GAGA_bind 136 pieeaaeeakekkkkkkrqrakkpkekkakkkkkksekskkkvkkesaderskaekksidlvlngvslDestlPvPvCsCtGalrqC 222
+ +++++++ + kk+ r++ +k +k+kk+kk +++k++v++ sa ++ k+ +ks+++v+ng+++D s++P+PvCsCtG+++qC
GSMUA_Achr11P09440_001 108 SVLPEPPTIEN-SPLKKTPRSSAQKPPKTKKPKKV-TAPKDEVANGSA-SHGKSGRKSTGMVINGINFDMSRIPTPVCSCTGKQQQC 191
33334444444.44555777888889999999994.567677777665.799*********************************** PP
GAGA_bind 223 YkWGnGGWqSaCCtttiSvyPLPvstkrrgaRiagrKmSqgafkklLekLaaeGydlsnpvDLkdhWAkHGtnkfvtir 301
Y+WG GGWqSaCCtt++S+yPLP+stkrrgaRiagrKmSqgafkk+LekLa+eGy+lsnp+DL+++WAkHGtnkfvtir
GSMUA_Achr11P09440_001 192 YRWGIGGWQSACCTTSVSIYPLPMSTKRRGARIAGRKMSQGAFKKVLEKLAGEGYNLSNPIDLRTFWAKHGTNKFVTIR 270
******************************************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01226 | 6.9E-127 | 1 | 270 | IPR010409 | GAGA-binding transcriptional activator |
| Pfam | PF06217 | 1.3E-96 | 1 | 270 | IPR010409 | GAGA-binding transcriptional activator |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0050793 | Biological Process | regulation of developmental process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 270 aa Download sequence Send to blast |
MDDDGGLGIR NWGYYEPSSK GNLGLRLMSS TVEQDAKPLL SNAGFIHRHC SIPEPTAPVD 60 FMSDGWFHHS NDNSKNDYVR DGLMRHSNNN SKNLHILPTS HQHHPTYSVL PEPPTIENSP 120 LKKTPRSSAQ KPPKTKKPKK VTAPKDEVAN GSASHGKSGR KSTGMVINGI NFDMSRIPTP 180 VCSCTGKQQQ CYRWGIGGWQ SACCTTSVSI YPLPMSTKRR GARIAGRKMS QGAFKKVLEK 240 LAGEGYNLSN PIDLRTFWAK HGTNKFVTIR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional regulator that specifically binds to GA-rich elements (GAGA-repeats) present in regulatory sequences of genes involved in developmental processes. Positively regulates the homeotic gene BKN3. {ECO:0000269|PubMed:12795701}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00253 | DAP | Transfer from AT2G01930 | Download |
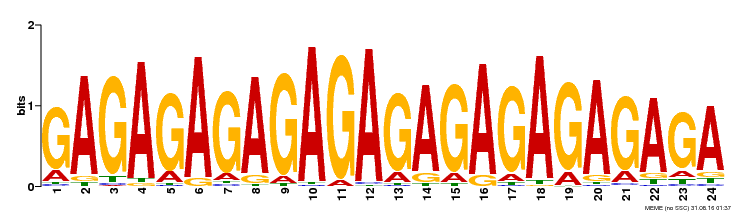
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009382984.1 | 0.0 | PREDICTED: protein Barley B recombinant-like isoform X2 | ||||
| Swissprot | Q8GUC3 | 1e-78 | BBR_HORVU; Protein Barley B recombinant | ||||
| TrEMBL | M0RQS4 | 0.0 | M0RQS4_MUSAM; Uncharacterized protein | ||||
| STRING | GSMUA_Achr11P09440_001 | 0.0 | (Musa acuminata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2211 | 36 | 97 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G01930.2 | 2e-71 | basic pentacysteine1 | ||||



