 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSMUA_Achr11P13410_001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Zingiberales; Musaceae; Musa
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 369aa MW: 39677.3 Da PI: 9.2497 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 101.2 | 6.6e-32 | 182 | 237 | 1 | 56 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRla 56
k+r++W+peLH+rF++a++qLGGs++AtPk+i+elmkv+gLt+++vkSHLQkYRl+
GSMUA_Achr11P13410_001 182 KARRCWSPELHQRFLHALKQLGGSHVATPKQIRELMKVDGLTNDEVKSHLQKYRLH 237
68****************************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 14.01 | 179 | 239 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.37E-17 | 180 | 239 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 4.0E-28 | 180 | 240 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 2.7E-26 | 182 | 237 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 5.5E-8 | 184 | 235 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 369 aa Download sequence Send to blast |
MDFSATLPSC GDYVRALEEE RKKIQVFQRE LPLCLQIVTH AIDSARRQMN GHATMSEDGP 60 ILEEFIPLKP SSSSSEDRSV DGAAKKSGSA ATRADEKPDW LRSVQLWNQE ADAFGKVEPP 120 TKPIAVNARR IGGAFHPFER EKHVAAPIPA SSSTTSGKGG VGGSGVGGEE KEKEGQSQPH 180 RKARRCWSPE LHQRFLHALK QLGGSHVATP KQIRELMKVD GLTNDEVKSH LQKYRLHNRR 240 PSTAVHSSSS NTTNPPIPQF VVVSGIWVPP PDYTVAAPPA DAPCTPAVGV FAPAASLPSD 300 SRVRPQQSKQ SDRSPTGPQL EKGGRSEESN SSGNAAAFNS LSPAASSSSQ TTTASHLEIT 360 CKSFMVGRQ |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 6e-14 | 182 | 240 | 2 | 60 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 6e-14 | 182 | 240 | 2 | 60 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_A | 6e-14 | 182 | 235 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 6e-14 | 182 | 235 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 6e-14 | 182 | 235 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 6e-14 | 182 | 235 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 6e-14 | 182 | 240 | 2 | 60 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 6e-14 | 182 | 240 | 2 | 60 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 6e-14 | 182 | 240 | 2 | 60 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 6e-14 | 182 | 240 | 2 | 60 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 6e-14 | 182 | 240 | 2 | 60 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 6e-14 | 182 | 240 | 2 | 60 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in phosphate signaling in roots. {ECO:0000250|UniProtKB:Q9FX67}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00160 | DAP | Transfer from AT1G25550 | Download |
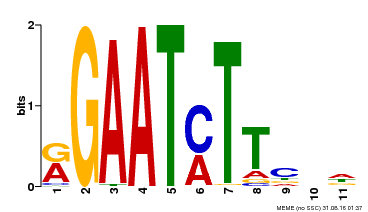
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009383634.1 | 0.0 | PREDICTED: myb family transcription factor EFM-like | ||||
| Swissprot | Q9FPE8 | 3e-90 | HHO3_ARATH; Transcription factor HHO3 | ||||
| TrEMBL | M0RRX1 | 0.0 | M0RRX1_MUSAM; Uncharacterized protein | ||||
| STRING | GSMUA_Achr11P13410_001 | 0.0 | (Musa acuminata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6470 | 37 | 54 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G25550.1 | 2e-80 | G2-like family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



