 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSMUA_Achr11P14570_001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Zingiberales; Musaceae; Musa
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 492aa MW: 54405.4 Da PI: 7.0478 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 29 | 2.4e-09 | 183 | 223 | 4 | 41 |
XCHHHCHHHHHHHHHHHHHHHHHHHHHHHH...HHHHHHHH CS
bZIP_1 4 lkrerrkqkNReAArrsRqRKkaeieeLee...kvkeLeae 41
k rr+++NReAAr+sR++Kka++++Le + k+Le+e
GSMUA_Achr11P14570_001 183 AKTLRRLAQNREAARKSRLKKKAYVQQLESsrlRLKQLEQE 223
6889*************************833344444444 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 2.8E-5 | 180 | 243 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 9.174 | 182 | 226 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 1.6E-6 | 183 | 223 | IPR004827 | Basic-leucine zipper domain |
| Gene3D | G3DSA:1.20.5.170 | 2.0E-7 | 183 | 225 | No hit | No description |
| SuperFamily | SSF57959 | 2.11E-6 | 184 | 227 | No hit | No description |
| CDD | cd14708 | 1.61E-22 | 184 | 235 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 187 | 202 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF14144 | 4.1E-29 | 266 | 340 | IPR025422 | Transcription factor TGA like domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 492 aa Download sequence Send to blast |
MATHRIGETG LSDSGPSTRS VSYGVHGTSL AASNFFDQEG AAYFGELEEA LMQGVDGIRE 60 IEDRKSFFGP RPATLEIFPS WPMRFQQTPR GKAHSARSTD SGSAQNTPSH PGSDSTSSRK 120 ETLMMMMTSG ASMTGTTPTL QPTSQDKISG NGNPSTLSIS LLFLLFLKMT DSTIEKHGKQ 180 LDAKTLRRLA QNREAARKSR LKKKAYVQQL ESSRLRLKQL EQELQRARSQ GLFLGVAGSS 240 NEAISPGAAM FDMEHARWLD ENCKLMSELR GALQAHLPEA SFTVIVDQCI MNYDELFQLK 300 AEVAKSDVFH LLSGSWTTPA ERSFLWMGGF RPSELLQILI PQLDPSTEQQ LIYNLKQSSQ 360 QAEEALSQGL EQLHQSLADT VAGGPLSDGV YVGNYMGHMV LALEKLAHLE GFVRQADNLR 420 QETLHQLRRI LTSRQAARCF VAIGEYYTRL RALSSLWASR PRQNLIGDDS VGSSTTDLQI 480 VHPSMQDHFS GF |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Required for the formation of the blade-sheath boundary in leaves. Promotes flowering. {ECO:0000269|PubMed:9490265}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00131 | DAP | Transfer from AT1G08320 | Download |
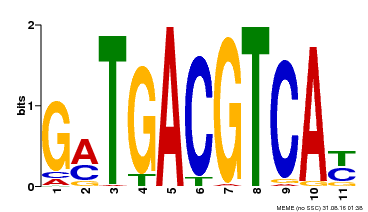
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009383673.2 | 0.0 | PREDICTED: transcription factor TGA2.2-like isoform X1 | ||||
| Swissprot | O49067 | 1e-169 | LG2_MAIZE; Transcription factor LG2 | ||||
| TrEMBL | M0RS87 | 0.0 | M0RS87_MUSAM; Uncharacterized protein | ||||
| STRING | GSMUA_Achr11P14570_001 | 0.0 | (Musa acuminata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3809 | 31 | 66 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G08320.3 | 1e-131 | bZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



